Genomic characterisation and ecological distribution of Mantoniella tinhauana: a novel Mamiellophycean green alga from the Western Pacific
- PMID: 38774501
- PMCID: PMC11106453
- DOI: 10.3389/fmicb.2024.1358574
Genomic characterisation and ecological distribution of Mantoniella tinhauana: a novel Mamiellophycean green alga from the Western Pacific
Abstract
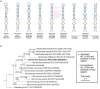
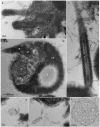
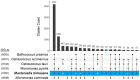
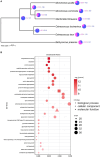
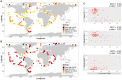
Mamiellophyceae are dominant marine algae in much of the ocean, the most prevalent genera belonging to the order Mamiellales: Micromonas, Ostreococcus and Bathycoccus, whose genetics and global distributions have been extensively studied. Conversely, the genus Mantoniella, despite its potential ecological importance, remains relatively under-characterised. In this study, we isolated and characterised a novel species of Mamiellophyceae, Mantoniella tinhauana, from subtropical coastal waters in the South China Sea. Morphologically, it resembles other Mantoniella species; however, a comparative analysis of the 18S and ITS2 marker genes revealed its genetic distinctiveness. Furthermore, we sequenced and assembled the first genome of Mantoniella tinhauana, uncovering significant differences from previously studied Mamiellophyceae species. Notably, the genome lacked any detectable outlier chromosomes and exhibited numerous unique orthogroups. We explored gene groups associated with meiosis, scale and flagella formation, shedding light on species divergence, yet further investigation is warranted. To elucidate the biogeography of Mantoniella tinhauana, we conducted a comprehensive analysis using global metagenomic read mapping to the newly sequenced genome. Our findings indicate this species exhibits a cosmopolitan distribution with a low-level prevalence worldwide. Understanding the intricate dynamics between Mamiellophyceae and the environment is crucial for comprehending their impact on the ocean ecosystem and accurately predicting their response to forthcoming environmental changes.
Keywords: Mamiellophyceae; Mantoniella tinhauana sp. nov.; biogeography; genomics; marine algae; metagenomics.
Copyright © 2024 Rey Redondo, Xu and Yung.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Aiyar A. (1999). “The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment” in Bioinformatics methods and protocols. eds. Misener S., Krawetz S. A. (Totowa, NJ: Humana Press; ).
-
- Alonso-González A., Orive E., David H., García-Etxebarria K., Garrido J. L., Laza-Martínez A., et al. (2014). Scaly green flagellates from Spanish Atlantic coastal waters: molecular, ultrastructural and pigment analyses. Bot. Mar. 57, 379–402. doi: 10.1515/bot-2013-0108 - DOI
-
- Baldwin B. G., Sanderson M. J., Porter J. M., Wojciechowski M. F., Campbell C. S., Donoghue M. J. (1995). The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 82, 247–277. doi: 10.2307/2399880 - DOI
-
- Balzano S., Gourvil P., Siano R., Chanoine M., Marie D., Lessard S., et al. (2012). Diversity of cultured photosynthetic flagellates in the north East Pacific and Arctic oceans in summer. Biogeosci. Discuss. 9, 4553–4571. doi: 10.5194/bgd-9-6219-2012 - DOI
-
- Bandi V., Gutwin C., Siri J. N., Neufeld E., Sharpe A., Parkin I. (2022). Visualization tools for genomic conservation. New York, NY: Springer US. - PubMed
LinkOut - more resources
Full Text Sources

