Advances in single-cell long-read sequencing technologies
- PMID: 38774511
- PMCID: PMC11106032
- DOI: 10.1093/nargab/lqae047
Advances in single-cell long-read sequencing technologies
Abstract
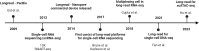
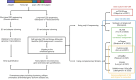
With an increase in accuracy and throughput of long-read sequencing technologies, they are rapidly being assimilated into the single-cell sequencing pipelines. For transcriptome sequencing, these techniques provide RNA isoform-level information in addition to the gene expression profiles. Long-read sequencing technologies not only help in uncovering complex patterns of cell-type specific splicing, but also offer unprecedented insights into the origin of cellular complexity and thus potentially new avenues for drug development. Additionally, single-cell long-read DNA sequencing enables high-quality assemblies, structural variant detection, haplotype phasing, resolving high-complexity regions, and characterization of epigenetic modifications. Given that significant progress has primarily occurred in single-cell RNA isoform sequencing (scRiso-seq), this review will delve into these advancements in depth and highlight the practical considerations and operational challenges, particularly pertaining to downstream analysis. We also aim to offer a concise introduction to complementary technologies for single-cell sequencing of the genome, epigenome and epitranscriptome. We conclude by identifying certain key areas of innovation that may drive these technologies further and foster more widespread application in biomedical science.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

References
LinkOut - more resources
Full Text Sources

