Pleiotropic roles of LAMMER kinase, Lkh1 in stress responses and virulence of Cryptococcus neoformans
- PMID: 38774630
- PMCID: PMC11106425
- DOI: 10.3389/fcimb.2024.1369301
Pleiotropic roles of LAMMER kinase, Lkh1 in stress responses and virulence of Cryptococcus neoformans
Abstract
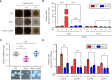
Dual-specificity LAMMER kinases are highly evolutionarily conserved in eukaryotes and play pivotal roles in diverse physiological processes, such as growth, differentiation, and stress responses. Although the functions of LAMMER kinase in fungal pathogens in pathogenicity and stress responses have been characterized, its role in Cryptococcus neoformans, a human fungal pathogen and a model yeast of basidiomycetes, remains elusive. In this study, we identified a LKH1 homologous gene and constructed a strain with a deleted LKH1 and a complemented strain. Similar to other fungi, the lkh1Δ mutant showed intrinsic growth defects. We observed that C. neoformans Lkh1 was involved in diverse stress responses, including oxidative stress and cell wall stress. Particularly, Lkh1 regulates DNA damage responses in Rad53-dependent and -independent manners. Furthermore, the absence of LKH1 reduced basidiospore formation. Our observations indicate that Lkh1 becomes hyperphosphorylated upon treatment with rapamycin, a TOR protein inhibitor. Notably, LKH1 deletion led to defects in melanin synthesis and capsule formation. Furthermore, we found that the deletion of LKH1 led to the avirulence of C. neoformans in a systemic cryptococcosis murine model. Taken together, Lkh1 is required for the stress response, sexual differentiation, and virulence of C. neoformans.
Keywords: Cryptococcus; LAMMER kinase; antifungal drug resistance; stress response; virulence.
Copyright © 2024 Kwon, Choi, Kim, Lee, Bahn and Jung.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures





Similar articles
-
Puf4 -mediated oxidative stress virulence attenuation in Cryptococcus neoformans.Front Cell Infect Microbiol. 2025 Aug 11;15:1628448. doi: 10.3389/fcimb.2025.1628448. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40861481 Free PMC article.
-
A flucytosine-responsive Mbp1/Swi4-like protein, Mbs1, plays pleiotropic roles in antifungal drug resistance, stress response, and virulence of Cryptococcus neoformans.Eukaryot Cell. 2012 Jan;11(1):53-67. doi: 10.1128/EC.05236-11. Epub 2011 Nov 11. Eukaryot Cell. 2012. PMID: 22080454 Free PMC article.
-
Regulatory Mechanism of the Atypical AP-1-Like Transcription Factor Yap1 in Cryptococcus neoformans.mSphere. 2019 Nov 20;4(6):e00785-19. doi: 10.1128/mSphere.00785-19. mSphere. 2019. PMID: 31748248 Free PMC article.
-
Stress-Activated Protein Kinases in Human Fungal Pathogens.Front Cell Infect Microbiol. 2019 Jul 17;9:261. doi: 10.3389/fcimb.2019.00261. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380304 Free PMC article. Review.
-
Cryptococcus neoformans: a sugar-coated killer with designer genes.FEMS Immunol Med Microbiol. 2005 Sep 1;45(3):395-404. doi: 10.1016/j.femsim.2005.06.005. FEMS Immunol Med Microbiol. 2005. PMID: 16055314 Review.
References
-
- Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., Lodge J. K. (2005). A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans . Eukaryot. Cell 4, 1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005 - DOI - PMC - PubMed
-
- Chang M., Sionov E., Khanal Lamichhane A., Kwon-Chung K. J., Chang Y. C. (2018). Roles of three Cryptococcus neoformans and Cryptococcus gattii efflux pump-coding genes in response to drug treatment. Antimicrob. Agents Chemother. 62, 10.1128/Aac. 01751–17. doi: 10.1128/AAC.01751-17 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

