CD4+ T cell heterogeneity in gestational age and preeclampsia using single-cell RNA sequencing
- PMID: 38774869
- PMCID: PMC11106458
- DOI: 10.3389/fimmu.2024.1401738
CD4+ T cell heterogeneity in gestational age and preeclampsia using single-cell RNA sequencing
Abstract
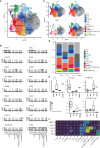
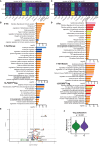
A balance between pro-inflammatory decidual CD4+ T cells and FOXP3+ regulatory T cells (FOXP3+ Tregs) is important for maintaining fetomaternal tolerance. Using single-cell RNA-sequencing and T cell receptor repertoire analysis, we determined that diversity and clonality of decidual CD4+ T cell subsets depend on gestational age. Th1/Th2 intermediate and Th1 subsets of CD4+ T cells were clonally expanded in both early and late gestation, whereas FOXP3+ Tregs were clonally expanded in late gestation. Th1/Th2 intermediate and FOXP3+ Treg subsets showed altered gene expression in preeclampsia (PE) compared to healthy late gestation. The Th1/Th2 intermediate subset exhibited elevated levels of cytotoxicity-related gene expression in PE. Moreover, increased Treg exhaustion was observed in the PE group, and FOXP3+ Treg subcluster analysis revealed that the effector Treg like subset drove the Treg exhaustion signatures in PE. The Th1/Th2 intermediate and effector Treg like subsets are possible inflammation-driving subsets in PE.
Keywords: CD4 + T cell; FOXP3; T cell receptor; immune tolerance; preeclampsia; pregnancy; regulatory T cell; single-cell RNA-sequencing.
Copyright © 2024 Tsuda, Shichino, Tilburgs, Shima, Morita, Yamaki-Ushijima, Roskin, Tomura, Sameshima, Saito and Nakashima.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

