Field-based high-throughput phenotyping enhances phenomic and genomic predictions for grain yield and plant height across years in maize
- PMID: 38776257
- PMCID: PMC11228873
- DOI: 10.1093/g3journal/jkae092
Field-based high-throughput phenotyping enhances phenomic and genomic predictions for grain yield and plant height across years in maize
Abstract
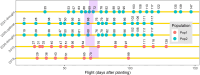
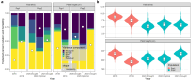
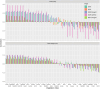
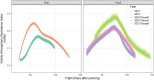
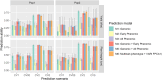
Field-based phenomic prediction employs novel features, like vegetation indices (VIs) from drone images, to predict key agronomic traits in maize, despite challenges in matching biomarker measurement time points across years or environments. This study utilized functional principal component analysis (FPCA) to summarize the variation of temporal VIs, uniquely allowing the integration of this data into phenomic prediction models tested across multiple years (2018-2021) and environments. The models, which included 1 genomic, 2 phenomic, 2 multikernel, and 1 multitrait type, were evaluated in 4 prediction scenarios (CV2, CV1, CV0, and CV00), relevant for plant breeding programs, assessing both tested and untested genotypes in observed and unobserved environments. Two hybrid populations (415 and 220 hybrids) demonstrated the visible atmospherically resistant index's strong temporal correlation with grain yield (up to 0.59) and plant height. The first 2 FPCAs explained 59.3 ± 13.9% and 74.2 ± 9.0% of the temporal variation of temporal data of VIs, respectively, facilitating predictions where flight times varied. Phenomic data, particularly when combined with genomic data, often were comparable to or numerically exceeded the base genomic model in prediction accuracy, particularly for grain yield in untested hybrids, although no significant differences in these models' performance were consistently observed. Overall, this approach underscores the effectiveness of FPCA and combined models in enhancing the prediction of grain yield and plant height across environments and diverse agricultural settings.
Keywords: UAV; field-based high-throughput phenotyping; functional principal component analysis; genomic prediction; grain yield; maize breeding; multikernel prediction; phenomic prediction; plant height.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures

Similar articles
-
Near-infrared reflectance spectroscopy phenomic prediction can perform similarly to genomic prediction of maize agronomic traits across environments.Plant Genome. 2024 Jun;17(2):e20454. doi: 10.1002/tpg2.20454. Epub 2024 May 7. Plant Genome. 2024. PMID: 38715204
-
Phenomic data-driven biological prediction of maize through field-based high-throughput phenotyping integration with genomic data.J Exp Bot. 2023 Sep 13;74(17):5307-5326. doi: 10.1093/jxb/erad216. J Exp Bot. 2023. PMID: 37279568
-
Unraveling the potential of phenomic selection within and among diverse breeding material of maize (Zea mays L.).G3 (Bethesda). 2022 Mar 4;12(3):jkab445. doi: 10.1093/g3journal/jkab445. G3 (Bethesda). 2022. PMID: 35100379 Free PMC article.
-
Integrating phenomic selection using single-kernel near-infrared spectroscopy and genomic selection for corn breeding improvement.Theor Appl Genet. 2025 Feb 26;138(3):60. doi: 10.1007/s00122-025-04843-w. Theor Appl Genet. 2025. PMID: 40009111 Free PMC article. Review.
-
Genomics-Assisted Breeding for Quantitative Disease Resistances in Small-Grain Cereals and Maize.Int J Mol Sci. 2020 Dec 19;21(24):9717. doi: 10.3390/ijms21249717. Int J Mol Sci. 2020. PMID: 33352763 Free PMC article. Review.
Cited by
-
110 years of rice breeding at LSU: realized genetic gains and future optimization.Theor Appl Genet. 2025 Jun 9;138(7):142. doi: 10.1007/s00122-025-04913-z. Theor Appl Genet. 2025. PMID: 40488752 Free PMC article.
-
Artificial Intelligence-Assisted Breeding for Plant Disease Resistance.Int J Mol Sci. 2025 Jun 1;26(11):5324. doi: 10.3390/ijms26115324. Int J Mol Sci. 2025. PMID: 40508136 Free PMC article. Review.
-
Using phenomic selection to predict hybrid values with NIR spectra measured on the parental lines: proof of concept on maize.Theor Appl Genet. 2025 Jan 11;138(1):28. doi: 10.1007/s00122-024-04809-4. Theor Appl Genet. 2025. PMID: 39797978 Free PMC article.
References
-
- Adak A, Anderson SL, Murray SC. 2023. Pedigree–management–flight interaction for temporal phenotype analysis and temporal phenomic prediction. Plant Phenome J. 6(1):e20057. doi:10.1002/ppj2.20057. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
