The Rac pathway prevents cell fragmentation in a nonprotrusively migrating leader cell during C. elegans gonad organogenesis
- PMID: 38776905
- PMCID: PMC12013728
- DOI: 10.1016/j.cub.2024.04.073
The Rac pathway prevents cell fragmentation in a nonprotrusively migrating leader cell during C. elegans gonad organogenesis
Abstract
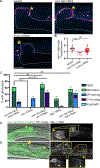
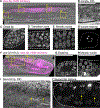
The C. elegans hermaphrodite distal tip cell (DTC) leads gonadogenesis. Loss-of-function mutations in a C. elegans ortholog of the Rac1 GTPase (ced-10) and its GEF complex (ced-5/DOCK180, ced-2/CrkII, ced-12/ELMO) cause gonad migration defects related to directional sensing; we discovered an additional defect class of gonad bifurcation in these mutants. Using genetic approaches, tissue-specific and whole-body RNAi, and in vivo imaging of endogenously tagged proteins and marked cells, we find that loss of Rac1 or its regulators causes the DTC to fragment as it migrates. Both products of fragmentation-the now-smaller DTC and the membranous patch of cellular material-localize important stem cell niche signaling (LAG-2 ligand) and migration (INA-1/integrin subunit alpha) factors to their membranes, but only one retains the DTC nucleus and therefore the ability to maintain gene expression over time. The enucleate patch can lead a bifurcating branch off the gonad arm that grows through germ cell proliferation. Germ cells in this branch differentiate as the patch loses LAG-2 expression. While the nucleus is surprisingly dispensable for aspects of leader cell function, it is required for stem cell niche activity long term. Prior work found that Rac1-/-;Rac2-/- mouse erythrocytes fragment; in this context, our new findings support the conclusion that maintaining a cohesive but deformable cell is a conserved function of this important cytoskeletal regulator.
Keywords: GTPase; Rac; fragmentation; germline; gonad; leader cell; migration; stem cell niche.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Getting there in one piece: The Rac pathway prevents cell fragmentation in a nonprotrusively migrating leader cell during organogenesis.bioRxiv [Preprint]. 2023 Dec 4:2023.12.01.569642. doi: 10.1101/2023.12.01.569642. bioRxiv. 2023. Update in: Curr Biol. 2024 Jun 3;34(11):2387-2402.e5. doi: 10.1016/j.cub.2024.04.073. PMID: 38106045 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

