Single nuclei RNA-seq reveals a medium spiny neuron glutamate excitotoxicity signature prior to the onset of neuronal death in an ovine Huntington's disease model
- PMID: 38776957
- PMCID: PMC11336116
- DOI: 10.1093/hmg/ddae087
Single nuclei RNA-seq reveals a medium spiny neuron glutamate excitotoxicity signature prior to the onset of neuronal death in an ovine Huntington's disease model
Abstract
Huntington's disease (HD) is a neurodegenerative genetic disorder caused by an expansion in the CAG repeat tract of the huntingtin (HTT) gene resulting in behavioural, cognitive, and motor defects. Current knowledge of disease pathogenesis remains incomplete, and no disease course-modifying interventions are in clinical use. We have previously reported the development and characterisation of the OVT73 transgenic sheep model of HD. The 73 polyglutamine repeat is somatically stable and therefore likely captures a prodromal phase of the disease with an absence of motor symptomatology even at 5-years of age and no detectable striatal cell loss. To better understand the disease-initiating events we have undertaken a single nuclei transcriptome study of the striatum of an extensively studied cohort of 5-year-old OVT73 HD sheep and age matched wild-type controls. We have identified transcriptional upregulation of genes encoding N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate receptors in medium spiny neurons, the cell type preferentially lost early in HD. Further, we observed an upregulation of astrocytic glutamate uptake transporters and medium spiny neuron GABAA receptors, which may maintain glutamate homeostasis. Taken together, these observations support the glutamate excitotoxicity hypothesis as an early neurodegeneration cascade-initiating process but the threshold of toxicity may be regulated by several protective mechanisms. Addressing this biochemical defect early may prevent neuronal loss and avoid the more complex secondary consequences precipitated by cell death.
Keywords: Huntington's disease; glutamate excitotoxicity; prodromal; single nuclei RNA-seq.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
J.F.G. was a founder and scientific advisory board member with a financial interest in Triplet Therapeutics Inc. His NIH-funded project is using genetic and genomic approaches to uncover other genes that significantly influence when diagnosable symptoms emerge and how rapidly they worsen in Huntington’s disease. The company was developing new therapeutic approaches to address triplet repeat disorders such Huntington’s disease, myotonic dystrophy and spinocerebellar ataxias. His interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. J.F.G. has also been a consultant for Wave Life Sciences USA Inc., Biogen Inc. and Pfizer Inc.
All other authors have declared that they have no competing interests.
Figures

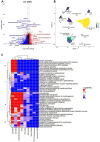
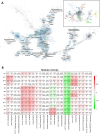
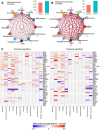

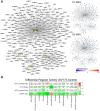
References
-
- The Huntington's Disease Collaborative Research Group . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's disease collaborative research group. Cell 1993;72:971–983. - PubMed
-
- Snell RG, MacMillan JC, Cheadle JP. et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet 1993;4:393–397. - PubMed
-
- Doble A. The role of excitotoxicity in neurodegenerative disease: implications for therapy. Pharmacol Ther 1999;81:163–221. - PubMed
-
- Salinska E, Danysz W, Lazarewicz JW. The role of excitotoxicity in neurodegeneration. Folia Neuropathol 2005;43:322–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

