Integrating human endogenous retroviruses into transcriptome-wide association studies highlights novel risk factors for major psychiatric conditions
- PMID: 38778015
- PMCID: PMC11111684
- DOI: 10.1038/s41467-024-48153-z
Integrating human endogenous retroviruses into transcriptome-wide association studies highlights novel risk factors for major psychiatric conditions
Abstract
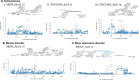
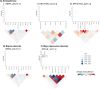
Human endogenous retroviruses (HERVs) are repetitive elements previously implicated in major psychiatric conditions, but their role in aetiology remains unclear. Here, we perform specialised transcriptome-wide association studies that consider HERV expression quantified to precise genomic locations, using RNA sequencing and genetic data from 792 post-mortem brain samples. In Europeans, we identify 1238 HERVs with expression regulated in cis, of which 26 represent expression signals associated with psychiatric disorders, with ten being conditionally independent from neighbouring expression signals. Of these, five are additionally significant in fine-mapping analyses and thus are considered high confidence risk HERVs. These include two HERV expression signatures specific to schizophrenia risk, one shared between schizophrenia and bipolar disorder, and one specific to major depressive disorder. No robust signatures are identified for autism spectrum conditions or attention deficit hyperactivity disorder in Europeans, or for any psychiatric trait in other ancestries, although this is likely a result of relatively limited statistical power. Ultimately, our study highlights extensive HERV expression and regulation in the adult cortex, including in association with psychiatric disorder risk, therefore providing a rationale for exploring neurological HERV expression in complex neuropsychiatric traits.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
- R01 MH109897/MH/NIMH NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 MH075916/MH/NIMH NIH HHS/United States
- U01 MH103392/MH/NIMH NIH HHS/United States
- R01 MH109677/MH/NIMH NIH HHS/United States
- R21 HG011513/HG/NHGRI NIH HHS/United States
- P50 MH084053/MH/NIMH NIH HHS/United States
- HHSN271201300031C/DA/NIDA NIH HHS/United States
- MR/W028018/1/MRC_/Medical Research Council/United Kingdom
- R01 MH110921/MH/NIMH NIH HHS/United States
- P50 MH066392/MH/NIMH NIH HHS/United States
- R01 MH080405/MH/NIMH NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
- R01 MH097276/MH/NIMH NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- R01 MH093725/MH/NIMH NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States

