From Planning Stage Towards FAIR Data: A Practical Metadatasheet For Biomedical Scientists
- PMID: 38778016
- PMCID: PMC11111677
- DOI: 10.1038/s41597-024-03349-2
From Planning Stage Towards FAIR Data: A Practical Metadatasheet For Biomedical Scientists
Abstract
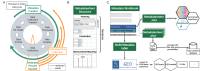
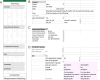
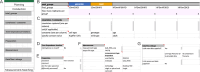
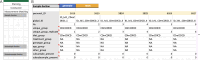
Datasets consist of measurement data and metadata. Metadata provides context, essential for understanding and (re-)using data. Various metadata standards exist for different methods, systems and contexts. However, relevant information resides at differing stages across the data-lifecycle. Often, this information is defined and standardized only at publication stage, which can lead to data loss and workload increase. In this study, we developed Metadatasheet, a metadata standard based on interviews with members of two biomedical consortia and systematic screening of data repositories. It aligns with the data-lifecycle allowing synchronous metadata recording within Microsoft Excel, a widespread data recording software. Additionally, we provide an implementation, the Metadata Workbook, that offers user-friendly features like automation, dynamic adaption, metadata integrity checks, and export options for various metadata standards. By design and due to its extensive documentation, the proposed metadata standard simplifies recording and structuring of metadata for biomedical scientists, promoting practicality and convenience in data management. This framework can accelerate scientific progress by enhancing collaboration and knowledge transfer throughout the intermediate steps of data creation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Morillo F, Bordons M, Gómez I. Interdisciplinarity in science: A tentative typology of disciplines and research areas. Journal of the American Society for Information Science and Technology. 2003;54:1237–1249. doi: 10.1002/asi.10326. - DOI
-
- Cioffi M, Goldman J, Marchese S. 2023. Harvard biomedical research data lifecycle. Zenodo. - DOI
-
- Habermann, T. Metadata life cycles, use cases and hierarchies. Geosciences8, 10.3390/geosciences8050179 (2018).
-
- Shaw F, et al. Copo: a metadata platform for brokering fair data in the life sciences. F1000Research. 2020;9:495. doi: 10.12688/f1000research.23889.1. - DOI
MeSH terms
Grants and funding
- 390685813/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 458597554/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- FE 1159/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- FE 1159/5-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- E 1159/2-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 432325352/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 450149205/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

