RUNX1-induced upregulation of PTGS2 enhances cell growth, migration and invasion in colorectal cancer cells
- PMID: 38778047
- PMCID: PMC11111780
- DOI: 10.1038/s41598-024-60296-z
RUNX1-induced upregulation of PTGS2 enhances cell growth, migration and invasion in colorectal cancer cells
Abstract
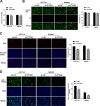
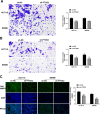
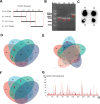
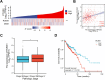
Colorectal cancer (CRC) arises via the progressive accumulation of dysregulation in key genes including oncogenes and tumor-suppressor genes. Prostaglandin-endoperoxide synthase 2 (PTGS2, also called COX2) acts as an oncogenic driver in CRC. Here, we explored the upstream transcription factors (TFs) responsible for elevating PTGS2 expression in CRC cells. The results showed that PTGS2 silencing repressed cell growth, migration and invasion in HCT116 and SW480 CRC cells. The two fragments (499-981 bp) and (1053-1434 bp) were confirmed as the core TF binding profiles of the PTGS2 promoter. PTGS2 expression positively correlated with RUNX1 level in colon adenocarcinoma (COAD) samples using the TCGA-COAD dataset. Furthermore, RUNX1 acted as a positive regulator of PTGS2 expression by promoting transcriptional activation of the PTGS2 promoter via the 1086-1096 bp binding motif. In conclusion, our study demonstrates that PTGS2 upregulation induced by the TF RUNX1 promotes CRC cell growth, migration and invasion, providing an increased rationale for the use of PTGS2 inhibitors in CRC prevention and treatment.
Keywords: Colorectal cancer; PTGS2; RUNX1; Transcription factor (TF).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
RUNX1 promotes colorectal cancer progression by activating SERPINE1 transcription.Biochem Biophys Res Commun. 2025 May 26;760:151732. doi: 10.1016/j.bbrc.2025.151732. Epub 2025 Mar 28. Biochem Biophys Res Commun. 2025. PMID: 40164015
-
RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer.J Exp Clin Cancer Res. 2019 Aug 1;38(1):334. doi: 10.1186/s13046-019-1330-9. J Exp Clin Cancer Res. 2019. PMID: 31370857 Free PMC article.
-
Prostaglandins induce early growth response 1 transcription factor mediated microsomal prostaglandin E2 synthase up-regulation for colorectal cancer progression.Oncotarget. 2015 Nov 24;6(37):39941-59. doi: 10.18632/oncotarget.5402. Oncotarget. 2015. PMID: 26498686 Free PMC article.
-
CD147 receptor is essential for TFF3-mediated signaling regulating colorectal cancer progression.Signal Transduct Target Ther. 2021 Jul 14;6(1):268. doi: 10.1038/s41392-021-00677-2. Signal Transduct Target Ther. 2021. PMID: 34262017 Free PMC article.
-
RUNX1-activated upregulation of lncRNA RNCR3 promotes cell proliferation, invasion, and suppresses apoptosis in colorectal cancer via miR-1301-3p/AKT1 axis in vitro and in vivo.Clin Transl Oncol. 2020 Oct;22(10):1762-1777. doi: 10.1007/s12094-020-02335-5. Epub 2020 Apr 1. Clin Transl Oncol. 2020. PMID: 32239427
Cited by
-
Comparison of the Efficacy of Photodynamic Therapy Versus Cisplatin Application.J Lasers Med Sci. 2024 Dec 24;15:e67. doi: 10.34172/jlms.2024.67. eCollection 2024. J Lasers Med Sci. 2024. PMID: 39949474 Free PMC article.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2021;71(3):209–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

