Selective haematological cancer eradication with preserved haematopoiesis
- PMID: 38778101
- PMCID: PMC11186773
- DOI: 10.1038/s41586-024-07456-3
Selective haematological cancer eradication with preserved haematopoiesis
Abstract
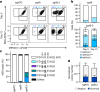
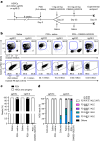
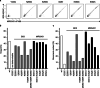
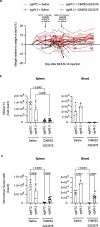
Haematopoietic stem cell (HSC) transplantation (HSCT) is the only curative treatment for a broad range of haematological malignancies, but the standard of care relies on untargeted chemotherapies and limited possibilities to treat malignant cells after HSCT without affecting the transplanted healthy cells1. Antigen-specific cell-depleting therapies hold the promise of much more targeted elimination of diseased cells, as witnessed in the past decade by the revolution of clinical practice for B cell malignancies2. However, target selection is complex and limited to antigens expressed on subsets of haematopoietic cells, resulting in a fragmented therapy landscape with high development costs2-5. Here we demonstrate that an antibody-drug conjugate (ADC) targeting the pan-haematopoietic marker CD45 enables the antigen-specific depletion of the entire haematopoietic system, including HSCs. Pairing this ADC with the transplantation of human HSCs engineered to be shielded from the CD45-targeting ADC enables the selective eradication of leukaemic cells with preserved haematopoiesis. The combination of CD45-targeting ADCs and engineered HSCs creates an almost universal strategy to replace a diseased haematopoietic system, irrespective of disease aetiology or originating cell type. We propose that this approach could have broad implications beyond haematological malignancies.
© 2024. The Author(s).
Conflict of interest statement
L.T.J. has a sponsored research agreement with Cimeio Therapeutics; the decision to publish was the sole responsibility of L.T.J., whose employer, Basel University Hospital, receives financial compensation for consulting work by L.T.J. I.D., L.C.W and C.K. are employed at Ridgeline Discovery. R.L., A.C. and S.U. are employed at Cimeio Therapeutics. P.H.v.B. is and I.K. was employed at ADC Therapeutics. L.T.J. is a co-founder and board member of Cimeio Therapeutics AG. The University of Basel, R.L., T.B., A.C., I.D., L.C.W., C.K., S.U. and L.T.J. hold equity in Cimeio Therapeutics. P.H.v.B. and I.K. hold equity in ADC Therapeutics. S.G., R.M., R.L., A.D., A.C., I.D., S.U. and L.T.J. are listed as inventors on a patent application related to the findings reported here. C.R.L. and S.Q.T. are inventors on a patent covering CHANGE-seq. S.Q.T. is a member of the scientific advisory board of Prime Medicine and Ensoma. L.T.J. receives: speaker fee from Novartis. F.S. receives: consulting fees from BMS/Celgene, Incyte and Kite/Gilead; speaker fees from Kite/Gilead and Incyte; and travel support from Kite/Gilead, Novartis, AstraZeneca, Neovii and Janssen. F.S. and F.B. receive research funding from Kite/Gilead, Novartis and BMS/Celgene. F.B. receives: institutional research funding from ADC Therapeutics, Bayer, BeiGene, Floratek Pharma, Helsinn, HTG Molecular Diagnostics, Ideogen, Idorsia Pharmaceuticals, Immagene, ImmunoGen, Menarini Ricerche, Nordic Nanovector, Oncternal Therapeutics and Spexis; consultancy fees from BIMINI Biotech, Helsinn and Menarini; advisory board fees to the institution from Novartis; and travel grants from Amgen, Astra Zeneca, Beigene and iOnctura. All other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

