PARP1-DOT1L transcription axis drives acquired resistance to PARP inhibitor in ovarian cancer
- PMID: 38778348
- PMCID: PMC11110363
- DOI: 10.1186/s12943-024-02025-8
PARP1-DOT1L transcription axis drives acquired resistance to PARP inhibitor in ovarian cancer
Abstract
Background: Poly (ADP-ribose) polymerase inhibitor (PARPi) resistance poses a significant challenge in ovarian carcinoma (OC). While the role of DOT1L in cancer and chemoresistance is acknowledged, its specific role in PARPi resistance remains unclear. This study aims to elucidate the molecular mechanism of DOT1L in PARPi resistance in OC patients.
Methods: This study analyzed the expression of DOT1L in PARPi-resistant cell lines compared to sensitive ones and correlated it with clinical outcomes in OC patients. Comprehensive in vitro and in vivo functional experiments were conducted using cellular and mouse models. Molecular investigations, including RNA sequencing, chromatin immunoprecipitation (ChIP) and Cleavage Under Targets and Tagmentation (CUT&Tag) assays, were employed to unravel the molecular mechanisms of DOT1L-mediated PARPi resistance.
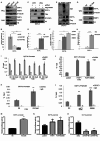
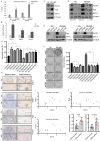
Results: Our investigation revealed a robust correlation between DOT1L expression and clinical PARPi resistance in non-BRCA mutated OC cells. Upregulated DOT1L expression in PARPi-resistant tissues was associated with diminished survival in OC patients. Mechanistically, we identified that PARP1 directly binds to the DOT1L gene promoter, promoting transcription independently of its enzyme activity. PARP1 trapping induced by PARPi treatment amplified this binding, enhancing DOT1L transcription and contributing to drug resistance. Sequencing analysis revealed that DOT1L plays a crucial role in the transcriptional regulation of PLCG2 and ABCB1 via H3K79me2. This established the PARP1-DOT1L-PLCG2/ABCB1 axis as a key contributor to PARPi resistance. Furthermore, we discovered that combining a DOT1L inhibitor with PARPi demonstrated a synergistic effect in both cell line-derived xenograft mouse models (CDXs) and patient-derived organoids (PDOs).
Conclusions: Our results demonstrate that DOT1L is an independent prognostic marker for OC patients. The PARP1-DOT1L/H3K79me2-PLCG2/ABCB1 axis is identified as a pivotal contributor to PARPi resistance. Targeted inhibition of DOT1L emerges as a promising therapeutic strategy for enhancing PARPi treatment outcomes in OC patients.
Keywords: DOT1L; Ovarian cancer; PARP1; PARPi resistance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
No potential conflicts of interest were disclosed.
Figures




References
-
- Coleman RL. Secondary Surgical cytoreduction for recurrent ovarian Cancer. Reply. N Engl J Med. 2020;382:686. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

