Heterotrophic bacteria trigger transcriptome remodelling in the photosynthetic picoeukaryote Micromonas commoda
- PMID: 38778545
- PMCID: PMC11112143
- DOI: 10.1111/1758-2229.13285
Heterotrophic bacteria trigger transcriptome remodelling in the photosynthetic picoeukaryote Micromonas commoda
Abstract
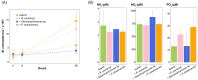
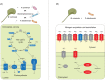
Marine biogeochemical cycles are built on interactions between surface ocean microbes, particularly those connecting phytoplankton primary producers to heterotrophic bacteria. Details of these associations are not well understood, especially in the case of direct influences of bacteria on phytoplankton physiology. Here we catalogue how the presence of three marine bacteria (Ruegeria pomeroyi DSS-3, Stenotrophomonas sp. SKA14 and Polaribacter dokdonensis MED152) individually and uniquely impact gene expression of the picoeukaryotic alga Micromonas commoda RCC 299. We find a dramatic transcriptomic remodelling by M. commoda after 8 h in co-culture, followed by an increase in cell numbers by 56 h compared with the axenic cultures. Some aspects of the algal transcriptomic response are conserved across all three bacterial co-cultures, including an unexpected reduction in relative expression of photosynthesis and carbon fixation pathways. Expression differences restricted to a single bacterium are also observed, with the Flavobacteriia P. dokdonensis uniquely eliciting changes in relative expression of algal genes involved in biotin biosynthesis and the acquisition and assimilation of nitrogen. This study reveals that M. commoda has rapid and extensive responses to heterotrophic bacteria in ways that are generalizable, as well as in a taxon specific manner, with implications for the diversity of phytoplankton-bacteria interactions ongoing in the surface ocean.
© 2024 The Author(s). Environmental Microbiology Reports published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Bacterial transcriptional response to labile exometabolites from photosynthetic picoeukaryote Micromonas commoda.ISME Commun. 2023 Jan 23;3(1):5. doi: 10.1038/s43705-023-00212-0. ISME Commun. 2023. PMID: 36690682 Free PMC article.
-
Epiphytic Bacteria Are Essential for the Production and Transformation of Algae-Derived Carboxyl-Rich Alicyclic Molecule (CRAM)-like DOM.Microbiol Spectr. 2021 Oct 31;9(2):e0153121. doi: 10.1128/Spectrum.01531-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668747 Free PMC article.
-
Sulfonate-based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean.Nat Microbiol. 2019 Oct;4(10):1706-1715. doi: 10.1038/s41564-019-0507-5. Epub 2019 Jul 22. Nat Microbiol. 2019. PMID: 31332382
-
Mini-review: Phytoplankton-derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria.Environ Microbiol. 2018 Aug;20(8):2671-2685. doi: 10.1111/1462-2920.14302. Epub 2018 Sep 9. Environ Microbiol. 2018. PMID: 30028074 Review.
-
Genomic insights into photosynthesis in eukaryotic phytoplankton.Trends Plant Sci. 2010 Oct;15(10):565-72. doi: 10.1016/j.tplants.2010.07.004. Epub 2010 Aug 26. Trends Plant Sci. 2010. PMID: 20800533 Review.
Cited by
-
Photosynthetic dependence and filament production in physical bacterial-Symbiodiniaceae interactions.ISME Commun. 2025 Apr 25;5(1):ycaf070. doi: 10.1093/ismeco/ycaf070. eCollection 2025 Jan. ISME Commun. 2025. PMID: 40385307 Free PMC article.
-
Laser-modulated MnBi2Te4 terahertz biosensor for high-sensitivity marine bacterial detection.Anal Bioanal Chem. 2025 Sep;417(21):4791-4801. doi: 10.1007/s00216-025-05996-9. Epub 2025 Aug 1. Anal Bioanal Chem. 2025. PMID: 40748359
References
-
- Amin, S.A. , Hmelo, L.R. , Van Tol, H.M. , Durham, B.P. , Carlson, L.T. , Heal, K.R. et al. (2015) Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature, 522, 98–101. - PubMed
-
- Armbrust, E.V. (2009) The life of diatoms in the world's oceans. Nature, 459, 185–192. - PubMed
-
- Armbrust, E.V. , Berges, J.A. , Bowler, C. , Green, B.R. , Martinez, D. , Putnam, N.H. et al. (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science, 306, 79–86. - PubMed
-
- Armbrust, E.V. , Parker, M.S. , Rocap, G. , Jenkins, B. & Bates, S. (2011) Pseudo‐nitzschia multiseries CLN‐47 draft genome assembly. https://mycocosm.jgi.doe.gov/Psemu1/Psemu1.home.html
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

