Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
- PMID: 38779102
- PMCID: PMC11110542
- DOI: 10.1007/978-3-031-43895-0_61
Longitudinal Multimodal Transformer Integrating Imaging and Latent Clinical Signatures From Routine EHRs for Pulmonary Nodule Classification
Abstract
The accuracy of predictive models for solitary pulmonary nodule (SPN) diagnosis can be greatly increased by incorporating repeat imaging and medical context, such as electronic health records (EHRs). However, clinically routine modalities such as imaging and diagnostic codes can be asynchronous and irregularly sampled over different time scales which are obstacles to longitudinal multimodal learning. In this work, we propose a transformer-based multimodal strategy to integrate repeat imaging with longitudinal clinical signatures from routinely collected EHRs for SPN classification. We perform unsupervised disentanglement of latent clinical signatures and leverage time-distance scaled self-attention to jointly learn from clinical signatures expressions and chest computed tomography (CT) scans. Our classifier is pretrained on 2,668 scans from a public dataset and 1,149 subjects with longitudinal chest CTs, billing codes, medications, and laboratory tests from EHRs of our home institution. Evaluation on 227 subjects with challenging SPNs revealed a significant AUC improvement over a longitudinal multimodal baseline (0.824 vs 0.752 AUC), as well as improvements over a single cross-section multimodal scenario (0.809 AUC) and a longitudinal imaging-only scenario (0.741 AUC). This work demonstrates significant advantages with a novel approach for co-learning longitudinal imaging and non-imaging phenotypes with transformers. Code available at https://github.com/MASILab/lmsignatures.
Keywords: Latent Clinical Signatures; Multimodal Transformers; Pulmonary Nodule Classification.
Figures
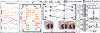
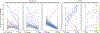

References
-
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G, et al.: End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography (May 2019), https://www.nature.com/articles/s41591-019-0447-x - PubMed
-
- Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W: Retain: An interpretable predictive model for healthcare using reverse time attention mechanism. Advances in neural information processing systems 29 (2016)
-
- Fritsch FN, Butland J: A method for constructing local monotone piecewise cubic interpolants. SIAM journal on scientific and statistical computing 5(2), 300–304 (1984)
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
