ULK1 Mediated Autophagy-Promoting Effects of Rutin-Loaded Chitosan Nanoparticles Contribute to the Activation of NF-κB Signaling Besides Inhibiting EMT in Hep3B Hepatoma Cells
- PMID: 38779103
- PMCID: PMC11110815
- DOI: 10.2147/IJN.S443117
ULK1 Mediated Autophagy-Promoting Effects of Rutin-Loaded Chitosan Nanoparticles Contribute to the Activation of NF-κB Signaling Besides Inhibiting EMT in Hep3B Hepatoma Cells
Abstract
Background: Liver cancer remains to be one of the leading causes of cancer worldwide. The treatment options face several challenges and nanomaterials have proven to improve the bioavailability of several drug candidates and their applications in nanomedicine. Specifically, chitosan nanoparticles (CNPs) are extremely biodegradable, pose enhanced biocompatibility and are considered safe for use in medicine.
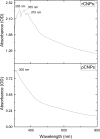
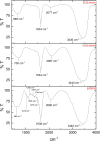
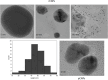
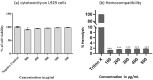
Methods: CNPs were synthesized by ionic gelation, loaded with rutin (rCNPs) and characterized by ultraviolet-visible spectroscopy (UV-Vis), Fourier-transform infrared spectroscopy (FTIR), dynamic light scattering (DLS) and transmission electron microscopy (TEM). The rCNPs were tested for their cytotoxic effects on human hepatoma Hep3B cells, and experiments were conducted to determine the mechanism of such effects. Further, the biocompatibility of the rCNPs was tested on L929 fibroblasts, and their hemocompatibility was determined.
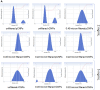
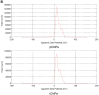
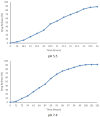
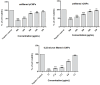
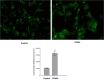
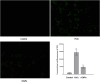
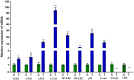
Results: Initially, UV-vis and FTIR analyses indicated the possible loading of rutin on rCNPs. Further, the rutin load was quantitatively measured using Ultra-Performance Liquid Chromatography (UPLC) and the concentration was 88 µg/mL for 0.22 micron filtered rCNPs. The drug loading capacity (LC%) of the rCNPs was observed to be 13.29 ± 0.68%, and encapsulation efficiency (EE%) was 19.55 ± 1.01%. The drug release was pH-responsive as 88.58% of the drug was released after 24 hrs at the lysosomal pH 5.5, whereas 91.44% of the drug was released at physiological pH 7.4 after 102 hrs. The cytotoxic effects were prominent in 0.22 micron filtered samples of 5 mg/mL rutin precursor. The particle size for the rCNPs at this concentration was 144.1 nm and the polydispersity index (PDI) was 0.244, which is deemed to be ideal for tumor targeting. A zeta potential (ζ-potential) value of 16.4 mV indicated rCNPs with good stability. The IC50 value for the cytotoxic effects of rCNPs on human hepatoma Hep3B cells was 9.7 ± 0.19 μg/mL of rutin load. In addition, the increased production of reactive oxygen species (ROS) and changes in mitochondrial membrane potential (MMP) were observed. Gene expression studies indicated that the mechanism for cytotoxic effects of rCNPs on Hep3B cells was due to the activation of Unc-51-like autophagy-activating kinase (ULK1) mediated autophagy and nuclear factor kappa B (NF-κB) signaling besides inhibiting the epithelial-mesenchymal Transition (EMT). In addition, the rCNPs were less toxic on NCTC clone 929 (L929) fibroblasts in comparison to the Hep3B cells and possessed excellent hemocompatibility (less than 2% of hemolysis).
Conclusion: The synthesized rCNPs were pH-responsive and possessed the physicochemical properties suitable for tumor targeting. The particles were effectively cytotoxic on Hep3B cells in comparison to normal cells and possessed excellent hemocompatibility. The very low hemolytic profile of rCNPs indicates that the drug could be administered intravenously for cancer therapy.
Keywords: Hep3B; ROS; mechanism; qPCR; rCNPs.
© 2024 Wu et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

