Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle
- PMID: 38779502
- PMCID: PMC11110911
- DOI: 10.3389/fmicb.2024.1386319
Bacterial enrichment prior to third-generation metagenomic sequencing improves detection of BRD pathogens and genetic determinants of antimicrobial resistance in feedlot cattle
Abstract
Introduction: Bovine respiratory disease (BRD) is one of the most important animal health problems in the beef industry. While bacterial culture and antimicrobial susceptibility testing have been used for diagnostic testing, the common practice of examining one isolate per species does not fully reflect the bacterial population in the sample. In contrast, a recent study with metagenomic sequencing of nasal swabs from feedlot cattle is promising in terms of bacterial pathogen identification and detection of antimicrobial resistance genes (ARGs). However, the sensitivity of metagenomic sequencing was impeded by the high proportion of host biomass in the nasal swab samples.
Methods: This pilot study employed a non-selective bacterial enrichment step before nucleic acid extraction to increase the relative proportion of bacterial DNA for sequencing.
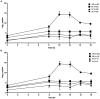
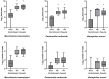
Results: Non-selective bacterial enrichment increased the proportion of bacteria relative to host sequence data, allowing increased detection of BRD pathogens compared with unenriched samples. This process also allowed for enhanced detection of ARGs with species-level resolution, including detection of ARGs for bacterial species of interest that were not targeted for culture and susceptibility testing. The long-read sequencing approach enabled ARG detection on individual bacterial reads without the need for assembly. Metagenomics following non-selective bacterial enrichment resulted in substantial agreement for four of six comparisons with culture for respiratory bacteria and substantial or better correlation with qPCR. Comparison between isolate susceptibility results and detection of ARGs was best for macrolide ARGs in Mannheimia haemolytica reads but was also substantial for sulfonamide ARGs within M. haemolytica and Pasteurella multocida reads and tetracycline ARGs in Histophilus somni reads.
Discussion: By increasing the proportion of bacterial DNA relative to host DNA through non-selective enrichment, we demonstrated a corresponding increase in the proportion of sequencing data identifying BRD-associated pathogens and ARGs in deep nasopharyngeal swabs from feedlot cattle using long-read metagenomic sequencing. This method shows promise as a detection strategy for BRD pathogens and ARGs and strikes a balance between processing time, input costs, and generation of on-target data. This approach could serve as a valuable tool to inform antimicrobial management for BRD and support antimicrobial stewardship.
Keywords: antimicrobial resistance; antimicrobial resistance genes; bovine respiratory disease; feedlot cattle; long-read metagenomic sequencing.
Copyright © 2024 Herman, Lacoste, Freeman, Otto, McCarthy, Links, Stothard and Waldner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
References
-
- Adewusi O. O., Waldner C. L., Hanington P. C., Hill J. E., Freeman C. N., Otto S. J. G. (2024). Laboratory tools for the direct detection of bacterial respiratory infections and antimicrobial resistance: a scoping review. J. Vet. Diagn. Invest.:10406387241235968. doi: 10.1177/10406387241235968, PMID: - DOI - PMC - PubMed
-
- Alhamami T., Chowdhury P., Gomes N., Carr M., Veltman T., Khazandi M., et al. (2021). First emergence of resistance to macrolides and tetracycline identified in Mannheimia haemolytica and Pasteurella multocida isolates from beef feedlots in Australia. Microorganisms 9:1322. doi: 10.3390/microorganisms9061322, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

