Extracellular host DNA contributes to pathogenic biofilm formation during periodontitis
- PMID: 38779563
- PMCID: PMC11109387
- DOI: 10.3389/fcimb.2024.1374817
Extracellular host DNA contributes to pathogenic biofilm formation during periodontitis
Abstract
Introduction: Periodontal diseases are known to be associated with polymicrobial biofilms and inflammasome activation. A deeper understanding of the subgingival cytological (micro) landscape, the role of extracellular DNA (eDNA) during periodontitis, and contribution of the host immune eDNA to inflammasome persistence, may improve our understanding of the mechanisms underlaying severe forms of periodontitis.
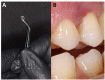
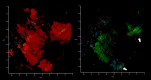
Methods: In this work, subgingival biolfilms developing on biologically neutral polyethylene terephthalate films placed in gingival cavities of patients with chronic periodontitis were investigated by confocal laser scanning microscopy (CLSM). This allowed examination of realistic cytological landscapes and visualization of extracellular polymeric substances (EPS) including amyloids, total proteins, carbohydrates and eDNA, as well as comparison with several single-strain in vitro model biofilms produced by oral pathogens such as Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus gordonii, S. sanguinis and S. mitis. Fluorescence in situ hybridization (FISH) analysis was also used to identify eDNA derived from eubacteria, streptococci and members of the Bacteroides-Porphyromonas-Prevotella (BPP) group associated with periodontitis.
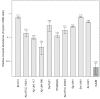
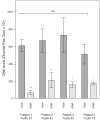
Results: Analysis of subgingival biofilm EPS revealed low levels of amyloids and high levels of eDNA which appears to be the main matrix component. However, bacterial eDNA contributed less than a third of the total eDNA observed, suggesting that host-derived eDNA released in neutrophil extracellular traps may be of more importance in the development of biofilms causing periodontitis.
Discussion: eDNA derived from host immunocompetent cells activated at the onset of periodontitis may therefore be a major driver of bacterial persistence and pathogenesis.
Keywords: FISH; biofilm structure; eDNA; periodontitis; subgingival biofilms.
Copyright © 2024 Slobodianyk-Kolomoiets, Khlebas, Mazur, Rudnieva, Potochilova, Iungin, Kamyshnyi, Kamyshna, Potters, Spiers and Moshynets.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









Similar articles
-
Identification of Extracellular DNA-Binding Proteins in the Biofilm Matrix.mBio. 2019 Jun 25;10(3):e01137-19. doi: 10.1128/mBio.01137-19. mBio. 2019. PMID: 31239382 Free PMC article.
-
Targeting the HUβ Protein Prevents Porphyromonas gingivalis from Entering into Preexisting Biofilms.J Bacteriol. 2018 May 9;200(11):e00790-17. doi: 10.1128/JB.00790-17. Print 2018 Jun 1. J Bacteriol. 2018. PMID: 29437850 Free PMC article.
-
Extracellular DNA in Helicobacter pylori biofilm: a backstairs rumour.J Appl Microbiol. 2011 Feb;110(2):490-8. doi: 10.1111/j.1365-2672.2010.04911.x. Epub 2010 Dec 10. J Appl Microbiol. 2011. PMID: 21143715
-
Unraveling the multifaceted role of extracellular DNA (eDNA) of biofilm in bacterial physiology, biofilm formation, and matrixome architecture.Crit Rev Biochem Mol Biol. 2025 Feb-Jun;60(1-3):1-32. doi: 10.1080/10409238.2025.2497270. Epub 2025 May 5. Crit Rev Biochem Mol Biol. 2025. PMID: 40322923 Review.
-
Subgingival biofilm structure.Front Oral Biol. 2012;15:1-16. doi: 10.1159/000329667. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142954 Review.
Cited by
-
Microbiome composition and metabolic pathways in shallow and deep periodontal pockets.Sci Rep. 2025 Apr 15;15(1):12926. doi: 10.1038/s41598-025-97531-0. Sci Rep. 2025. PMID: 40234709 Free PMC article.
-
Clarithromycin Modulates Neutrophilic Inflammation Induced by Prevotella intermedia in Human Airway Epithelial Cells.Antibiotics (Basel). 2024 Sep 23;13(9):909. doi: 10.3390/antibiotics13090909. Antibiotics (Basel). 2024. PMID: 39335081 Free PMC article.
-
Dental biofilms contain DNase I-resistant Z-DNA and G-quadruplexes but alternative DNase overcomes this resistance.NPJ Biofilms Microbiomes. 2025 May 19;11(1):80. doi: 10.1038/s41522-025-00694-x. NPJ Biofilms Microbiomes. 2025. PMID: 40389511 Free PMC article.
References
-
- Amann R. I., Binder B. J., Olson R. J., Chisholm S. W., Devereux R., Stahl D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

