An integrative multi-omic analysis defines gut microbiota, mycobiota, and metabolic fingerprints in ulcerative colitis patients
- PMID: 38779566
- PMCID: PMC11109417
- DOI: 10.3389/fcimb.2024.1366192
An integrative multi-omic analysis defines gut microbiota, mycobiota, and metabolic fingerprints in ulcerative colitis patients
Abstract
Background: Ulcerative colitis (UC) is a multifactorial chronic inflammatory bowel disease (IBD) that affects the large intestine with superficial mucosal inflammation. A dysbiotic gut microbial profile has been associated with UC. Our study aimed to characterize the UC gut bacterial, fungal, and metabolic fingerprints by omic approaches.
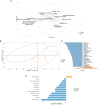
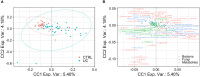
Methods: The 16S rRNA- and ITS2-based metataxonomics and gas chromatography-mass spectrometry/solid phase microextraction (GC-MS/SPME) metabolomic analysis were performed on stool samples of 53 UC patients and 37 healthy subjects (CTRL). Univariate and multivariate approaches were applied to separated and integrated omic data, to define microbiota, mycobiota, and metabolic signatures in UC. The interaction between gut bacteria and fungi was investigated by network analysis.
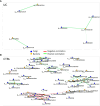
Results: In the UC cohort, we reported the increase of Streptococcus, Bifidobacterium, Enterobacteriaceae, TM7-3, Granulicatella, Peptostreptococcus, Lactobacillus, Veillonella, Enterococcus, Peptoniphilus, Gemellaceae, and phenylethyl alcohol; and we also reported the decrease of Akkermansia; Ruminococcaceae; Ruminococcus; Gemmiger; Methanobrevibacter; Oscillospira; Coprococus; Christensenellaceae; Clavispora; Vishniacozyma; Quambalaria; hexadecane; cyclopentadecane; 5-hepten-2-ol, 6 methyl; 3-carene; caryophyllene; p-Cresol; 2-butenal; indole, 3-methyl-; 6-methyl-3,5-heptadiene-2-one; 5-octadecene; and 5-hepten-2-one, 6 methyl. The integration of the multi-omic data confirmed the presence of a distinctive bacterial, fungal, and metabolic fingerprint in UC gut microbiota. Moreover, the network analysis highlighted bacterial and fungal synergistic and/or divergent interkingdom interactions.
Conclusion: In this study, we identified intestinal bacterial, fungal, and metabolic UC-associated biomarkers. Furthermore, evidence on the relationships between bacterial and fungal ecosystems provides a comprehensive perspective on intestinal dysbiosis and ecological interactions between microorganisms in the framework of UC.
Keywords: dysbiosis; gut metabolism; gut microbiota; inflammatory bowel disease; intestinal biomarkers; multi-omic integrated approaches; ulcerative colitis.
Copyright © 2024 Scanu, Toto, Petito, Masi, Fidaleo, Puca, Baldelli, Reddel, Vernocchi, Pani, Putignani, Scaldaferri and Del Chierico.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

