Kcnma1 alternative splicing in mouse kidney: regulation during development and by dietary K+ intake
- PMID: 38779757
- PMCID: PMC12151499
- DOI: 10.1152/ajprenal.00100.2024
Kcnma1 alternative splicing in mouse kidney: regulation during development and by dietary K+ intake
Abstract
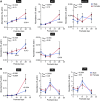
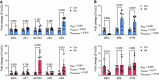
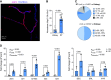
The pore-forming α-subunit of the large-conductance K+ (BK) channel is encoded by a single gene, KCNMA1. BK channel-mediated K+ secretion in the kidney is crucial for overall renal K+ homeostasis in both physiological and pathological conditions. BK channels achieve phenotypic diversity by various mechanisms, including substantial exon rearrangements at seven major alternative splicing sites. However, KCNMA1 alternative splicing in the kidney has not been characterized. The present study aims to identify the major splice variants of mouse Kcnma1 in whole kidney and distal nephron segments. We designed primers that specifically cross exons within each alternative splice site of mouse Kcnma1 and performed real-time quantitative RT-PCR (RT-qPCR) to quantify relative abundance of each splice variant. Our data suggest that Kcnma1 splice variants within mouse kidney are less diverse than in the brain. During postnatal kidney development, most Kcnma1 splice variants at site 5 and the COOH terminus increase in abundance over time. Within the kidney, the regulation of Kcnma1 alternative exon splicing within these two sites by dietary K+ loading is both site and sex specific. In microdissected distal tubules, the Kcnma1 alternative splicing profile, as well as its regulation by dietary K+, are distinctly different than in the whole kidney, suggesting segment and/or cell type specificity in Kcnma1 splicing events. Overall, our data provide evidence that Kcnma1 alternative splicing is regulated during postnatal development and may serve as an important adaptive mechanism to dietary K+ loading in mouse kidney.NEW & NOTEWORTHY We identified the major Kcnma1 splice variants that are specifically expressed in the whole mouse kidney or aldosterone-sensitive distal nephron segments. Our data suggest that Kcnma1 alternative splicing is developmentally regulated and subject to changes in dietary K+.
Keywords: BK channel; Kcnma1; kidney; potassium; splice variants.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures



Similar articles
-
Paraoxonase 3 regulates the pore-forming α subunit of the large-conductance K+ channel.Biochim Biophys Acta Mol Cell Res. 2025 Aug;1872(6):119990. doi: 10.1016/j.bbamcr.2025.119990. Epub 2025 May 12. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 40368269 Free PMC article.
-
Structural mapping of patient-associated KCNMA1 gene variants.Biophys J. 2024 Jul 16;123(14):1984-2000. doi: 10.1016/j.bpj.2023.11.3404. Epub 2023 Dec 1. Biophys J. 2024. PMID: 38042986 Free PMC article.
-
Conservation of alternative splicing in sodium channels reveals evolutionary focus on release from inactivation and structural insights into gating.J Physiol. 2017 Aug 15;595(16):5671-5685. doi: 10.1113/JP274693. Epub 2017 Jul 18. J Physiol. 2017. PMID: 28621020 Free PMC article.
-
Antiemetics for adults for prevention of nausea and vomiting caused by moderately or highly emetogenic chemotherapy: a network meta-analysis.Cochrane Database Syst Rev. 2021 Nov 16;11(11):CD012775. doi: 10.1002/14651858.CD012775.pub2. Cochrane Database Syst Rev. 2021. PMID: 34784425 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
Cited by
-
Going with the flow: New insights regarding flow induced K+ secretion in the distal nephron.Physiol Rep. 2024 Oct;12(20):e70087. doi: 10.14814/phy2.70087. Physiol Rep. 2024. PMID: 39428258 Free PMC article. Review.
-
Paraoxonase 3 regulates the pore-forming α subunit of the large-conductance K+ channel.Biochim Biophys Acta Mol Cell Res. 2025 Aug;1872(6):119990. doi: 10.1016/j.bbamcr.2025.119990. Epub 2025 May 12. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 40368269 Free PMC article.
-
Runs of homozygosity and selection signals analysis reveals domestication traits and divergence in local domestic duck breeds.Poult Sci. 2025 Jun 6;104(9):105404. doi: 10.1016/j.psj.2025.105404. Online ahead of print. Poult Sci. 2025. PMID: 40513394 Free PMC article.
References
-
- Knaus HG, Folander K, Garcia-Calvo M, Garcia ML, Kaczorowski GJ, Smith M, Swanson R. Primary sequence and immunological characterization of beta-subunit of high conductance Ca2+-activated K+ channel from smooth muscle. J Biol Chem 269: 17274–17278, 1994. - PubMed
MeSH terms
Substances
Grants and funding
- DK137329/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- DK130901/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- U54 DK137329/DK/NIDDK NIH HHS/United States
- R01 DK130901/DK/NIDDK NIH HHS/United States
- HL128053/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- HL158295/HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- DK038470/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK111380/DK/NIDDK NIH HHS/United States
- DK129285/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK038470/DK/NIDDK NIH HHS/United States
- T32 DK061296/DK/NIDDK NIH HHS/United States
- R01 HL128053/HL/NHLBI NIH HHS/United States
- T32 DK007052/DK/NIDDK NIH HHS/United States
- R56 DK111380/DK/NIDDK NIH HHS/United States
- R25 HL158295/HL/NHLBI NIH HHS/United States
- DK111380/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- DK061296/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- DK007052/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- R01 DK129285/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

