Large-scale mutational analysis identifies UNC93B1 variants that drive TLR-mediated autoimmunity in mice and humans
- PMID: 38780621
- PMCID: PMC11116816
- DOI: 10.1084/jem.20232005
Large-scale mutational analysis identifies UNC93B1 variants that drive TLR-mediated autoimmunity in mice and humans
Abstract
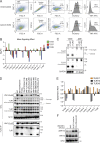
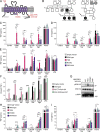
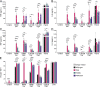
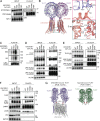
Nucleic acid-sensing Toll-like receptors (TLR) 3, 7/8, and 9 are key innate immune sensors whose activities must be tightly regulated to prevent systemic autoimmune or autoinflammatory disease or virus-associated immunopathology. Here, we report a systematic scanning-alanine mutagenesis screen of all cytosolic and luminal residues of the TLR chaperone protein UNC93B1, which identified both negative and positive regulatory regions affecting TLR3, TLR7, and TLR9 responses. We subsequently identified two families harboring heterozygous coding mutations in UNC93B1, UNC93B1+/T93I and UNC93B1+/R336C, both in key negative regulatory regions identified in our screen. These patients presented with cutaneous tumid lupus and juvenile idiopathic arthritis plus neuroinflammatory disease, respectively. Disruption of UNC93B1-mediated regulation by these mutations led to enhanced TLR7/8 responses, and both variants resulted in systemic autoimmune or inflammatory disease when introduced into mice via genome editing. Altogether, our results implicate the UNC93B1-TLR7/8 axis in human monogenic autoimmune diseases and provide a functional resource to assess the impact of yet-to-be-reported UNC93B1 mutations.
© 2024 Rael et al.
Conflict of interest statement
Disclosures: A.M. Stevens reported personal fees from Janssen Pharmaceuticals and personal fees from Acelyrin, Inc. outside the submitted work. G.M. Barton serves on the scientific advisory boards for Actym Therapeutics and X-Biotix and has provided consulting services for Lycia Therapeutics. No other disclosures were reported.
Figures





References
-
- Baum, R., Sharma S., Carpenter S., Li Q.Z., Busto P., Fitzgerald K.A., Marshak-Rothstein A., and Gravallese E.M.. 2015. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. J. Immunol. 194:873–877. 10.4049/jimmunol.1402573 - DOI - PMC - PubMed
-
- Biesen, R., Demir C., Barkhudarova F., Grün J.R., Steinbrich-Zöllner M., Backhaus M., Häupl T., Rudwaleit M., Riemekasten G., Radbruch A., et al. . 2008. Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum. 58:1136–1145. 10.1002/art.23404 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 1752814/National Science Foundation
- R01AI072429/NH/NIH HHS/United States
- Office of the NIH
- T32 AR079068/AR/NIAMS NIH HHS/United States
- U01 NS134356/NS/NINDS NIH HHS/United States
- Lupus Research Institute
- T32 AI100829/AI/NIAID NIH HHS/United States
- U01 NS134355/NS/NINDS NIH HHS/United States
- Children's Mercy Research Institute
- U01 IP001050/IP/NCIRD CDC HHS/United States
- U01IP001050/CC/CDC HHS/United States
- U01 HG007703/HG/NHGRI NIH HHS/United States
- Office of Strategic Coordination
- R01 AI072429/AI/NIAID NIH HHS/United States
- Chan-Zuckerburg Biohub Physician Scientist Fellowship Program
- HHMI/Howard Hughes Medical Institute/United States
- U01 HG010218/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

