Mitotic deacetylase complex (MiDAC) recognizes the HIV-1 core promoter to control activated viral gene expression
- PMID: 38781120
- PMCID: PMC11115230
- DOI: 10.1371/journal.ppat.1011821
Mitotic deacetylase complex (MiDAC) recognizes the HIV-1 core promoter to control activated viral gene expression
Abstract
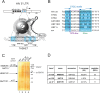
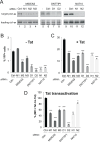
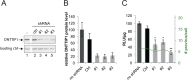
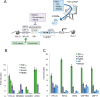
The human immunodeficiency virus (HIV) integrates into the host genome forming latent cellular reservoirs that are an obstacle for cure or remission strategies. Viral transcription is the first step in the control of latency and depends upon the hijacking of the host cell RNA polymerase II (Pol II) machinery by the 5' HIV LTR. Consequently, "block and lock" or "shock and kill" strategies for an HIV cure depend upon a full understanding of HIV transcriptional control. The HIV trans-activating protein, Tat, controls HIV latency as part of a positive feed-forward loop that strongly activates HIV transcription. The recognition of the TATA box and adjacent sequences of HIV essential for Tat trans-activation (TASHET) of the core promoter by host cell pre-initiation complexes of HIV (PICH) has been shown to be necessary for Tat trans-activation, yet the protein composition of PICH has remained obscure. Here, DNA-affinity chromatography was employed to identify the mitotic deacetylase complex (MiDAC) as selectively recognizing TASHET. Using biophysical techniques, we show that the MiDAC subunit DNTTIP1 binds directly to TASHET, in part via its CTGC DNA motifs. Using co-immunoprecipitation assays, we show that DNTTIP1 interacts with MiDAC subunits MIDEAS and HDAC1/2. The Tat-interacting protein, NAT10, is also present in HIV-bound MiDAC. Gene silencing revealed a functional role for DNTTIP1, MIDEAS, and NAT10 in HIV expression in cellulo. Furthermore, point mutations in TASHET that prevent DNTTIP1 binding block the reactivation of HIV by latency reversing agents (LRA) that act via the P-TEFb/7SK axis. Our data reveal a key role for MiDAC subunits DNTTIP1, MIDEAS, as well as NAT10, in Tat-activated HIV transcription and latency. DNTTIP1, MIDEAS and NAT10 emerge as cell cycle-regulated host cell transcription factors that can control activated HIV gene expression, and as new drug targets for HIV cure strategies.
Copyright: © 2024 Wilhelm et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
PPMcD is also employed as Executive Director, Research at Insmed Inc. Insmed was however not involved in this work; did not fund it; and does not endorse it, implicitly or otherwise. B.B. owns shares in the biotechnology company Ascioma. Ascioma had no involvement with this research, financial or otherwise.
Figures




References
-
- Blancou P, Chenciner N, Ho Tsong Fang R, Monceaux V, Cumont MC, Guetard D, et al.. Simian immunodeficiency virus promoter exchange results in a highly attenuated strain that protects against uncloned challenge virus. J Virol. 2004;78(3):1080–92. Epub 2004/01/15. doi: 10.1128/jvi.78.3.1080-1092.2004 ; PubMed Central PMCID: PMC321388. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

