IL-33 controls IL-22-dependent antibacterial defense by modulating the microbiota
- PMID: 38781213
- PMCID: PMC11145264
- DOI: 10.1073/pnas.2310864121
IL-33 controls IL-22-dependent antibacterial defense by modulating the microbiota
Abstract
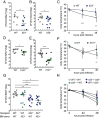
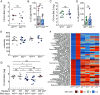
IL-22 plays a critical role in defending against mucosal infections, but how IL-22 production is regulated is incompletely understood. Here, we show that mice lacking IL-33 or its receptor ST2 (IL-1RL1) were more resistant to Streptococcus pneumoniae lung infection than wild-type animals and that single-nucleotide polymorphisms in IL33 and IL1RL1 were associated with pneumococcal pneumonia in humans. The effect of IL-33 on S. pneumoniae infection was mediated by negative regulation of IL-22 production in innate lymphoid cells (ILCs) but independent of ILC2s as well as IL-4 and IL-13 signaling. Moreover, IL-33's influence on IL-22-dependent antibacterial defense was dependent on housing conditions of the mice and mediated by IL-33's modulatory effect on the gut microbiota. Collectively, we provide insight into the bidirectional crosstalk between the innate immune system and the microbiota. We conclude that both genetic and environmental factors influence the gut microbiota, thereby impacting the efficacy of antibacterial immune defense and susceptibility to pneumonia.
Keywords: IL-22; IL-33; Streptococcus pneumoniae; microbiota; pneumonia.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



Comment in
-
Gut microbes coordinate pulmonary immunity.Proc Natl Acad Sci U S A. 2024 Jun 18;121(25):e2408800121. doi: 10.1073/pnas.2408800121. Epub 2024 Jun 6. Proc Natl Acad Sci U S A. 2024. PMID: 38843253 Free PMC article. No abstract available.
References
-
- Huijts S. M., et al. , Diagnostic accuracy of a serotype-specific antigen test in community-acquired pneumonia. Eur. Respir. J. 42, 1283–1290 (2013). - PubMed
-
- Thibeault C., Suttorp N., Opitz B., The microbiota in pneumonia: From protection to predisposition. Sci. Transl. Med. 13, eaba0501 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- SFB-TR84 A1/Deutsche Forschungsgemeinschaft (DFG)
- OP86/13-1/Deutsche Forschungsgemeinschaft (DFG)
- TRR 167 - 259373024/Deutsche Forschungsgemeinschaft (DFG)
- FOR2599 - 322359157/Deutsche Forschungsgemeinschaft (DFG)
- TRR 241 - 375876048/Deutsche Forschungsgemeinschaft (DFG)
- SPP1937 - KL 2963/2-1 and KL 2963/3-1/Deutsche Forschungsgemeinschaft (DFG)
- MAPVAP FKZ 01KI2124/Bundesministerium für Bildung und Forschung (BMBF)
- ERCEA; 803087/EC | ERC | HORIZON EUROPE European Research Council (ERC)
- SFB1444 TP11/Deutsche Forschungsgemeinschaft (DFG)
- Advanced Grant 101055309 - ILCADAPT/EC | ERC | HORIZON EUROPE European Research Council (ERC)
- Einstein Professor/Einstein Foundation Berlin
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

