Machine learning-enhanced molecular network reveals global exposure to hundreds of unknown PFAS
- PMID: 38781329
- PMCID: PMC11114235
- DOI: 10.1126/sciadv.adn1039
Machine learning-enhanced molecular network reveals global exposure to hundreds of unknown PFAS
Abstract
Unknown forever chemicals like per- and polyfluoroalkyl substances (PFASs) are difficult to identify. Current platforms designed for metabolites and natural products cannot capture the diverse structural characteristics of PFAS. Here, we report an automatic PFAS identification platform (APP-ID) that screens for PFAS in environmental samples using an enhanced molecular network and identifies unknown PFAS structures using machine learning. Our networking algorithm, which enhances characteristic fragment matches, has lower false-positive rate (0.7%) than current algorithms (2.4 to 46%). Our support vector machine model identified unknown PFAS in test set with 58.3% accuracy, surpassing current software. Further, APP-ID detected 733 PFASs in real fluorochemical wastewater, 39 of which are previously unreported in environmental media. Retrospective screening of 126 PFASs against public data repository from 20 countries show PFAS substitutes are prevalent worldwide.
Figures
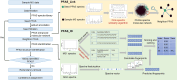


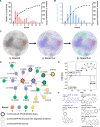

References
-
- Trang B., Li Y., Xue X.-S., Ateia M., Houk K. N., Dichtel W. R., Low-temperature mineralization of perfluorocarboxylic acids. Science 377, 839–845 (2022). - PubMed
-
- Evich M. G., Davis M. J. B., McCord J. P., Acrey B., Awkerman J. A., Knappe D. R. U., Lindstrom A. B., Speth T. F., Tebes-Stevens C., Strynar M. J., Wang Z., Weber E. J., Henderson W. M., Washington J. W., Per- and polyfluoroalkyl substances in the environment. Science 375, eabg9065 (2022). - PMC - PubMed
-
- Zhu J.-J., Dressel W., Pacion K., Ren Z. J., ES&T in the 21st century: A data-driven analysis of research topics, interconnections, and trends in the past 20 years. Environ. Sci. Technol. 55, 3453–3464 (2021). - PubMed
-
- Sen P., Qadri S., Luukkonen P. K., Ragnarsdottir O., McGlinchey A., Jäntti S., Juuti A., Arola J., Schlezinger J. J., Webster T. F., Orešič M., Yki-Järvinen H., Hyötyläinen T., Exposure to environmental contaminants is associated with altered hepatic lipid metabolism in non-alcoholic fatty liver disease. J. Hepatol. 76, 283–293 (2022). - PubMed
-
- Bartell S. M., Vieira V. M., Critical review on PFOA, kidney cancer, and testicular cancer. J. Air Waste Manage. Assoc. 71, 663–679 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

