Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface
- PMID: 38782204
- PMCID: PMC11237949
- DOI: 10.1016/j.jbc.2024.107404
Structure and dynamics of the staphylococcal pyridoxal 5-phosphate synthase complex reveal transient interactions at the enzyme interface
Abstract
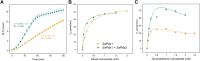
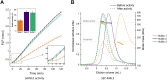
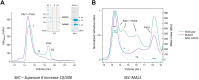
Infectious diseases are a significant cause of death, and recent studies estimate that common bacterial infectious diseases were responsible for 13.6% of all global deaths in 2019. Among the most significant bacterial pathogens is Staphylococcus aureus, accounting for more than 1.1 million deaths worldwide in 2019. Vitamin biosynthesis has been proposed as a promising target for antibacterial therapy. Here, we investigated the biochemical, structural, and dynamic properties of the enzyme complex responsible for vitamin B6 (pyridoxal 5-phosphate, PLP) biosynthesis in S. aureus, which comprises enzymes SaPdx1 and SaPdx2. The crystal structure of the 24-mer complex of SaPdx1-SaPdx2 enzymes indicated that the S. aureus PLP synthase complex forms a highly dynamic assembly with transient interaction between the enzymes. Solution scattering data indicated that SaPdx2 typically binds to SaPdx1 at a substoichiometric ratio. We propose a structure-based view of the PLP synthesis mechanism initiated with the assembly of SaPLP synthase complex that proceeds in a highly dynamic interaction between Pdx1 and Pdx2. This interface interaction can be further explored as a potentially druggable site for the design of new antibiotics.
Keywords: PLP synthase; Staphylococcus aureus; oligomeric state; protein crystallization; vitamin B6.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

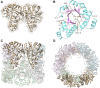

Similar articles
-
Dissection of contributions from invariant amino acids to complex formation and catalysis in the heteromeric pyridoxal 5-phosphate synthase complex from Bacillus subtilis.Biochemistry. 2009 Mar 10;48(9):1928-35. doi: 10.1021/bi801887r. Biochemistry. 2009. PMID: 19152323
-
Solution Structures and Dynamic Assembly of the 24-Meric Plasmodial Pdx1-Pdx2 Complex.Int J Mol Sci. 2020 Aug 19;21(17):5971. doi: 10.3390/ijms21175971. Int J Mol Sci. 2020. PMID: 32825141 Free PMC article.
-
Crystal structures capture three states in the catalytic cycle of a pyridoxal phosphate (PLP) synthase.J Biol Chem. 2015 Feb 27;290(9):5226-39. doi: 10.1074/jbc.M114.626382. Epub 2015 Jan 7. J Biol Chem. 2015. PMID: 25568319 Free PMC article.
-
Structural Dynamics and Perspectives of Vitamin B6 Biosynthesis Enzymes in Plasmodium: Advances and Open Questions.Front Cell Infect Microbiol. 2021 Jul 13;11:688380. doi: 10.3389/fcimb.2021.688380. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34327152 Free PMC article. Review.
-
Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein.Arch Biochem Biophys. 2005 Jan 1;433(1):144-56. doi: 10.1016/j.abb.2004.08.037. Arch Biochem Biophys. 2005. PMID: 15581573 Review.
Cited by
-
Structure and identification of the native PLP synthase complex from Methanosarcina acetivorans lysate.mBio. 2025 Jan 8;16(1):e0309024. doi: 10.1128/mbio.03090-24. Epub 2024 Nov 26. mBio. 2025. PMID: 39589128 Free PMC article.
References
-
- Lee A.S., De Lencastre H., Garau J., Al E. Methicillin resistance in Staphylococcus aureus. Nat. Rev. Dis. Primers. 2018;4:225–235. - PubMed
-
- Weiner-Lastinger L.M., Abner S., Edwards J.R., Kallen A.J., Karlsson M., Magill S.S., et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 2020;41:1–18. - PMC - PubMed
-
- Stefani S., Chung D.R., Lindsay J.A., Friedrich A.W., Kearns A.M., Westh H., et al. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents. 2012;39:273–282. - PubMed
-
- Khoshnood S., Heidary M., Asadi A., Soleimani S., Motahar M., Savari M., et al. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed. Pharmacother. 2019;109:1809–1818. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

