In Silico Assisted Identification, Synthesis, and In Vitro Pharmacological Characterization of Potent and Selective Blockers of the Epilepsy-Associated KCNT1 Channel
- PMID: 38782404
- PMCID: PMC11181338
- DOI: 10.1021/acs.jmedchem.4c00268
In Silico Assisted Identification, Synthesis, and In Vitro Pharmacological Characterization of Potent and Selective Blockers of the Epilepsy-Associated KCNT1 Channel
Abstract
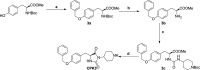
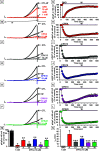
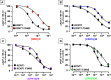
Gain-of-function (GoF) variants in KCNT1 channels cause severe, drug-resistant forms of epilepsy. Quinidine is a known KCNT1 blocker, but its clinical use is limited due to severe drawbacks. To identify novel KCNT1 blockers, a homology model of human KCNT1 was built and used to screen an in-house library of compounds. Among the 20 molecules selected, five (CPK4, 13, 16, 18, and 20) showed strong KCNT1-blocking ability in an in vitro fluorescence-based assay. Patch-clamp experiments confirmed a higher KCNT1-blocking potency of these compounds when compared to quinidine, and their selectivity for KCNT1 over hERG and Kv7.2 channels. Among identified molecules, CPK20 displayed the highest metabolic stability; this compound also blocked KCNT2 currents, although with a lower potency, and counteracted GoF effects prompted by 2 recurrent epilepsy-causing KCNT1 variants (G288S and A934T). The present results provide solid rational basis for future design of novel compounds to counteract KCNT1-related neurological disorders.
Conflict of interest statement
The authors declare no competing financial interest.
Figures











References
-
- Franceschetti S.; Lavazza T.; Curia G.; Aracri P.; Panzica F.; Sancini G.; Avanzini G.; Magistretti J. Na+-Activated K+ Current Contributes to Postexcitatory Hyperpolarization in Neocortical Intrinsically Bursting Neurons. J. Neurophysiol. 2003, 89 (4), 2101–2111. 10.1152/jn.00695.2002. - DOI - PubMed

