Chemokine receptor hetero-oligomers regulate monocyte chemotaxis
- PMID: 38782603
- PMCID: PMC11116815
- DOI: 10.26508/lsa.202402657
Chemokine receptor hetero-oligomers regulate monocyte chemotaxis
Abstract
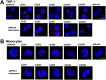
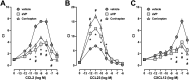
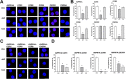
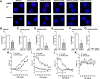
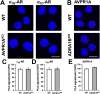
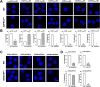
It is known that stress influences immune cell function. The underlying molecular mechanisms are unclear. We recently reported that many chemokine receptors (CRs) heteromerize with α1-adrenoceptors (α1-ARs) through which CRs are regulated. Here, we show that arginine vasopressin receptor 1A (AVPR1A) heteromerizes with all human CRs, except chemokine (C-X-C motif) receptor (CXCR)1, in recombinant systems and that such heteromers are detectable in THP-1 cells and human monocytes. We demonstrate that ligand-free AVPR1A differentially regulates the efficacy of CR partners to mediate chemotaxis and that AVPR1A ligands disrupt AVPR1A:CR heteromers, which enhances chemokine (C-C motif) receptor (CCR)1-mediated chemotaxis and inhibits CCR2-, CCR8-, and CXCR4-mediated chemotaxis. Using bioluminescence resonance energy transfer to monitor G protein activation and CRISPR/Cas9 gene-edited THP-1 cells lacking AVPR1A or α1B-AR, we show that CRs that share the propensity to heteromerize with α1B/D-ARs and AVPR1A exist and function within interdependent hetero-oligomeric complexes through which the efficacy of CRs to mediate chemotaxis is controlled. Our findings suggest that hetero-oligomers composed of CRs, α1B/D-ARs, and AVPR1A may enable stress hormones to regulate immune cell trafficking.
© 2024 Enten et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
