Epigenome-metabolism nexus in the retina: implications for aging and disease
- PMID: 38782642
- PMCID: PMC11303112
- DOI: 10.1016/j.tig.2024.04.012
Epigenome-metabolism nexus in the retina: implications for aging and disease
Abstract
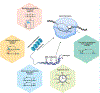
Intimate links between epigenome modifications and metabolites allude to a crucial role of cellular metabolism in transcriptional regulation. Retina, being a highly metabolic tissue, adapts by integrating inputs from genetic, epigenetic, and extracellular signals. Precise global epigenomic signatures guide development and homeostasis of the intricate retinal structure and function. Epigenomic and metabolic realignment are hallmarks of aging and highlight a link of the epigenome-metabolism nexus with aging-associated multifactorial traits affecting the retina, including age-related macular degeneration and glaucoma. Here, we focus on emerging principles of epigenomic and metabolic control of retinal gene regulation, with emphasis on their contribution to human disease. In addition, we discuss potential mitigation strategies involving lifestyle changes that target the epigenome-metabolome relationship for maintaining retinal function.
Keywords: DNA methylation; age-related macular degeneration; diet; environmental factors; gene regulation; histone modifications; metabolic pathways; multifactorial traits; vision impairment.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


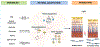
Similar articles
-
Epitranscriptome-epigenome interactions in development and disease mechanisms.Trends Genet. 2025 Aug;41(8):691-705. doi: 10.1016/j.tig.2025.04.009. Epub 2025 May 15. Trends Genet. 2025. PMID: 40374434 Review.
-
Eye on the horizon: The metabolic landscape of the RPE in aging and disease.Prog Retin Eye Res. 2025 Jan;104:101306. doi: 10.1016/j.preteyeres.2024.101306. Epub 2024 Oct 19. Prog Retin Eye Res. 2025. PMID: 39433211 Free PMC article. Review.
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
Epigenomic signatures of accelerated epigenetic aging are associated with congenital heart disease in newborns.BMC Med Genomics. 2025 Jul 28;18(1):121. doi: 10.1186/s12920-025-02189-2. BMC Med Genomics. 2025. PMID: 40722095 Free PMC article.
-
Secreted IgM deficiency alters the retinal landscape enhancing neurodegeneration associated with aging.Immun Ageing. 2025 Feb 24;22(1):9. doi: 10.1186/s12979-025-00502-2. Immun Ageing. 2025. PMID: 39994686 Free PMC article.
Cited by
-
DNA hypermethylation of PLTP mediated by DNMT3B aggravates vascular dysfunction in diabetic retinopathy via the AKT/GSK3β signaling pathway.Clin Epigenetics. 2025 May 17;17(1):82. doi: 10.1186/s13148-025-01874-4. Clin Epigenetics. 2025. PMID: 40380281 Free PMC article.
-
Age-Related Macular Degeneration: Cellular and Molecular Signaling Mechanisms.Int J Mol Sci. 2025 Jun 26;26(13):6174. doi: 10.3390/ijms26136174. Int J Mol Sci. 2025. PMID: 40649951 Free PMC article. Review.
-
Single-cell analysis of the epigenome and 3D chromatin architecture in the human retina.bioRxiv [Preprint]. 2025 Apr 2:2024.12.28.630634. doi: 10.1101/2024.12.28.630634. bioRxiv. 2025. PMID: 39764062 Free PMC article. Preprint.
-
Transcriptional Heterogeneity and Differential Response of Rod Photoreceptor Pathway Uncovered by Single-Cell RNA Sequencing of the Aging Mouse Retina.Aging Cell. 2025 May;24(5):e70001. doi: 10.1111/acel.70001. Epub 2025 Feb 15. Aging Cell. 2025. PMID: 39954235 Free PMC article.
-
Epigenetic Modifications in the Retinal Pigment Epithelium of the Eye During RPE-Related Regeneration or Retinal Diseases in Vertebrates.Biomedicines. 2025 Jun 25;13(7):1552. doi: 10.3390/biomedicines13071552. Biomedicines. 2025. PMID: 40722628 Free PMC article. Review.
References
-
- Jaenisch R and Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics 33 Suppl, 245–254 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

