Autophagy-deficient macrophages exacerbate cisplatin-induced mitochondrial dysfunction and kidney injury via miR-195a-5p-SIRT3 axis
- PMID: 38782909
- PMCID: PMC11116430
- DOI: 10.1038/s41467-024-47842-z
Autophagy-deficient macrophages exacerbate cisplatin-induced mitochondrial dysfunction and kidney injury via miR-195a-5p-SIRT3 axis
Abstract
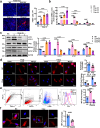
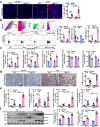
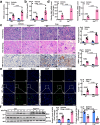
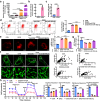
Macrophages (Mφ) autophagy is a pivotal contributor to inflammation-related diseases. However, the mechanistic details of its direct role in acute kidney injury (AKI) were unclear. Here, we show that Mφ promote AKI progression via crosstalk with tubular epithelial cells (TECs), and autophagy of Mφ was activated and then inhibited in cisplatin-induced AKI mice. Mφ-specific depletion of ATG7 (Atg7Δmye) aggravated kidney injury in AKI mice, which was associated with tubulointerstitial inflammation. Moreover, Mφ-derived exosomes from Atg7Δmye mice impaired TEC mitochondria in vitro, which may be attributable to miR-195a-5p enrichment in exosomes and its interaction with SIRT3 in TECs. Consistently, either miR-195a-5p inhibition or SIRT3 overexpression improved mitochondrial bioenergetics and renal function in vivo. Finally, adoptive transfer of Mφ from AKI mice to Mφ-depleted mice promotes the kidney injury response to cisplatin, which is alleviated when Mφ autophagy is activated with trehalose. We conclude that exosomal miR-195a-5p mediate the communication between autophagy-deficient Mφ and TECs, leading to impaired mitochondrial biogenetic in TECs and subsequent exacerbation of kidney injury in AKI mice via miR-195a-5p-SIRT3 axis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

