Reduction of specific enterocytes from loss of intestinal LGR4 improves lipid metabolism in mice
- PMID: 38782937
- PMCID: PMC11116434
- DOI: 10.1038/s41467-024-48622-5
Reduction of specific enterocytes from loss of intestinal LGR4 improves lipid metabolism in mice
Abstract
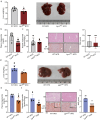
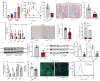
Whether intestinal Leucine-rich repeat containing G-protein-coupled receptor 4 (LGR4) impacts nutrition absorption and energy homeostasis remains unknown. Here, we report that deficiency of Lgr4 (Lgr4iKO) in intestinal epithelium decreased the proportion of enterocytes selective for long-chain fatty acid absorption, leading to reduction in lipid absorption and subsequent improvement in lipid and glucose metabolism. Single-cell RNA sequencing demonstrates the heterogeneity of absorptive enterocytes, with a decrease in enterocytes selective for long-chain fatty acid-absorption and an increase in enterocytes selective for carbohydrate absorption in Lgr4iKO mice. Activation of Notch signaling and concurrent inhibition of Wnt signaling are observed in the transgenes. Associated with these alterations is the substantial reduction in lipid absorption. Decrement in lipid absorption renders Lgr4iKO mice resistant to high fat diet-induced obesity relevant to wild type littermates. Our study thus suggests that targeting intestinal LGR4 is a potential strategy for the intervention of obesity and liver steatosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Luo CW, Hsueh AJ. Genomic analyses of the evolution of LGR genes. Chang. Gung. Med. J. 2006;29:2–8. - PubMed
MeSH terms
Substances
Grants and funding
- R01 DK110273/DK/NIDDK NIH HHS/United States
- 82330017/National Natural Science Foundation of China (National Science Foundation of China)
- 81930015/National Natural Science Foundation of China (National Science Foundation of China)
- R01 DK112755/DK/NIDDK NIH HHS/United States
- R01 DK129360/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

