naRNA-LL37 composite DAMPs define sterile NETs as self-propagating drivers of inflammation
- PMID: 38783164
- PMCID: PMC11239898
- DOI: 10.1038/s44319-024-00150-5
naRNA-LL37 composite DAMPs define sterile NETs as self-propagating drivers of inflammation
Abstract
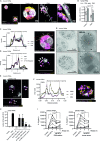
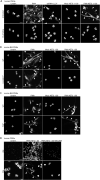
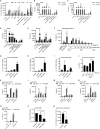
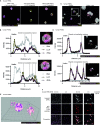
Neutrophil extracellular traps (NETs) are a key antimicrobial feature of cellular innate immunity mediated by polymorphonuclear neutrophils (PMNs). NETs counteract microbes but are also linked to inflammation in atherosclerosis, arthritis, or psoriasis by unknown mechanisms. Here, we report that NET-associated RNA (naRNA) stimulates further NET formation in naive PMNs via a unique TLR8-NLRP3 inflammasome-dependent pathway. Keratinocytes respond to naRNA with expression of psoriasis-related genes (e.g., IL17, IL36) via atypical NOD2-RIPK signaling. In vivo, naRNA drives temporary skin inflammation, which is drastically ameliorated by genetic ablation of RNA sensing. Unexpectedly, the naRNA-LL37 'composite damage-associated molecular pattern (DAMP)' is pre-stored in resting neutrophil granules, defining sterile NETs as inflammatory webs that amplify neutrophil activation. However, the activity of the naRNA-LL37 DAMP is transient and hence supposedly self-limiting under physiological conditions. Collectively, upon dysregulated NET release like in psoriasis, naRNA sensing may represent both a potential cause of disease and a new intervention target.
Keywords: DAMP; NLRP3 Inflammasome; Neutrophil Extracellular Trap; RNA; Toll-like Receptors.
© 2024. The Author(s).
Conflict of interest statement
NKA has received previous grant support from Pfizer and Boehringer Ingelheim and was a paid consultant for Janssen Pharmaceuticals. MWL is an inventor of patents owned by Immatics Biotechnologies and has acted as a speaker and paid consultant for Boehringer Ingelheim. All other authors declare no competing interests.
Figures


References
-
- Abtin A, Eckhart L, Mildner M, Ghannadan M, Harder J, Schroder JM, Tschachler E. Degradation by stratum corneum proteases prevents endogenous RNase inhibitor from blocking antimicrobial activities of RNase 5 and RNase 7. J Invest Dermatol. 2009;129:2193–2201. doi: 10.1038/jid.2009.35. - DOI - PubMed
-
- Balak DM, van Doorn MB, Arbeit RD, Rijneveld R, Klaassen E, Sullivan T, Brevard J, Thio HB, Prens EP, Burggraaf J, Rissmann R. IMO-8400, a toll-like receptor 7, 8, and 9 antagonist, demonstrates clinical activity in a phase 2a, randomized, placebo-controlled trial in patients with moderate-to-severe plaque psoriasis. Clin Immunol. 2017;174:63–72. doi: 10.1016/j.clim.2016.09.015. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R01 AR073665/AR/NIAMS NIH HHS/United States
- SCHI510/12-1/Deutsche Forschungsgemeinschaft (DFG)
- Momentum grant "InnatelyHuman"/Volkswagen Foundation (VolkswagenStiftung)
- We-4195/18-1/Deutsche Forschungsgemeinschaft (DFG)
- EXC-2180/Deutsche Forschungsgemeinschaft (DFG)
- EXC-2124/Deutsche Forschungsgemeinschaft (DFG)
- 2021/13049-4/Sao Paolo Research Foundation (FAPESP)
- R01 AR069502/AR/NIAMS NIH HHS/United States
- R01AI146177, R01AR073665, and R01AR069502/HHS | National Institutes of Health (NIH)
- INST 35/1259/Deutsche Forschungsgemeinschaft (DFG)
- R01 AI146177/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

