Multi-repeat sequences identification using genome mining techniques for developing highly sensitive molecular diagnostic assay for the detection of Chlamydia trachomatis
- PMID: 38783971
- PMCID: PMC11109563
- DOI: 10.12688/openresafrica.14316.2
Multi-repeat sequences identification using genome mining techniques for developing highly sensitive molecular diagnostic assay for the detection of Chlamydia trachomatis
Abstract
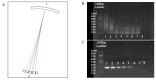
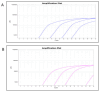
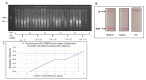
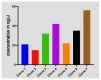
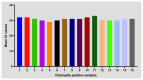
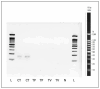
Chlamydia trachomatis ( C. trachomatis) is a common sexually transmitted infection (STI). In 2019, the World Health Organization reported about 131 million infections. The majority of infected patients are asymptomatic with cases remaining undetected. It is likely that missed C. trachomatis infections contribute to preventable adverse health outcomes in women and children. Consequently, there is an urgent need of developing efficient diagnostic methods. In this study, genome-mining approaches to identify identical multi-repeat sequences (IMRS) distributed throughout the C. trachomatis genome were used to design a primer pair that would target regions in the genome. Genomic DNA was 10-fold serially diluted (100pg/μL to 1×10 -3pg/μL) and used as DNA template for PCR reactions. The gold standard PCR using 16S rRNA primers was also run as a comparative test, and products were resolved on agarose gel. The novel assay, C. trachomatis IMRS-PCR, had an analytical sensitivity of 4.31 pg/µL, representing better sensitivity compared with 16S rRNA PCR (9.5 fg/µL). Our experimental data demonstrate the successful development of lateral flow and isothermal assays for detecting C. trachomatis DNA with potential use in field settings. There is a potential to implement this concept in miniaturized, isothermal, microfluidic platforms, and laboratory-on-a-chip diagnostic devices for reliable point-of-care testing.
Keywords: Chlamydia trachomatis; Identical Multi-Repeat Sequences; Isothermal assays.
Copyright: © 2024 Shiluli C et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
Improving gonorrhoea molecular diagnostics: Genome mining-based identification of identical multi-repeat sequences (IMRS) in Neisseria gonorrhoeae.Heliyon. 2024 Mar 5;10(6):e27344. doi: 10.1016/j.heliyon.2024.e27344. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38533083 Free PMC article.
-
Highly sensitive molecular assay based on Identical Multi-Repeat Sequence (IMRS) algorithm for the detection of Trichomonas vaginalis infection.PLoS One. 2025 Feb 7;20(2):e0317958. doi: 10.1371/journal.pone.0317958. eCollection 2025. PLoS One. 2025. PMID: 39919090 Free PMC article.
-
Prevalence of Chlamydia infection among women visiting a gynaecology outpatient department: evaluation of an in-house PCR assay for detection of Chlamydia trachomatis.Ann Clin Microbiol Antimicrob. 2010 Sep 8;9:24. doi: 10.1186/1476-0711-9-24. Ann Clin Microbiol Antimicrob. 2010. PMID: 20822551 Free PMC article.
-
Rapid point of care test for detecting urogenital Chlamydia trachomatis infection in nonpregnant women and men at reproductive age.Cochrane Database Syst Rev. 2020 Jan 29;1(1):CD011708. doi: 10.1002/14651858.CD011708.pub2. Cochrane Database Syst Rev. 2020. PMID: 31995238 Free PMC article.
-
DNA amplification assays: a new standard for diagnosis of Chlamydia trachomatis infections.Ann Acad Med Singap. 1995 Jul;24(4):627-33. Ann Acad Med Singap. 1995. PMID: 8849200 Review.
References
-
- Wynn A, Mussa A, Ryan R, et al. : Evaluating the diagnosis and treatment of Chlamydia trachomatis and Neisseria gonorrhoeae in pregnant women to prevent adverse neonatal consequences in Gaborone, Botswana: protocol for the Maduo study. BMC Infect Dis. 2022;22(1): 229. 10.1186/s12879-022-07093-z - DOI - PMC - PubMed
-
- World Health Organization: Sexually transmitted infections.2019;27:9–10. Reference Source
LinkOut - more resources
Full Text Sources

