Quantitative proteomics reveals the dynamic proteome landscape of zebrafish embryos during the maternal-to-zygotic transition
- PMID: 38784018
- PMCID: PMC11111832
- DOI: 10.1016/j.isci.2024.109944
Quantitative proteomics reveals the dynamic proteome landscape of zebrafish embryos during the maternal-to-zygotic transition
Abstract
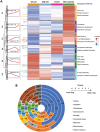
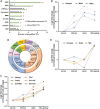
Maternal-to-zygotic transition (MZT) is central to early embryogenesis. However, its underlying molecular mechanisms are still not well described. Here, we revealed the expression dynamics of 5,000 proteins across four stages of zebrafish embryos during MZT, representing one of the most systematic surveys of proteome landscape of the zebrafish embryos during MZT. Nearly 700 proteins were differentially expressed and were divided into six clusters according to their expression patterns. The proteome expression profiles accurately reflect the main events that happen during the MZT, i.e., zygotic genome activation (ZGA), clearance of maternal mRNAs, and initiation of cellular differentiation and organogenesis. MZT is modulated by many proteins at multiple levels in a collaborative fashion, i.e., transcription factors, histones, histone-modifying enzymes, RNA helicases, and P-body proteins. Significant discrepancies were discovered between zebrafish proteome and transcriptome profiles during the MZT. The proteome dynamics database will be a valuable resource for bettering our understanding of MZT.
Keywords: Developmental biology; Proteomics.
© 2024 The Author(s).
Conflict of interest statement
Authors declare that they have no competing interests.
Figures




Similar articles
-
Dynamic RNA-protein interactions underlie the zebrafish maternal-to-zygotic transition.Genome Res. 2017 Jul;27(7):1184-1194. doi: 10.1101/gr.215954.116. Epub 2017 Apr 5. Genome Res. 2017. PMID: 28381614 Free PMC article.
-
SLAMseq resolves the kinetics of maternal and zygotic gene expression during early zebrafish embryogenesis.Cell Rep. 2023 Feb 28;42(2):112070. doi: 10.1016/j.celrep.2023.112070. Epub 2023 Feb 8. Cell Rep. 2023. PMID: 36757845
-
Comparative transcriptome analysis explores maternal to zygotic transition during Eriocheir sinensis early embryogenesis.Gene. 2019 Feb 15;685:12-20. doi: 10.1016/j.gene.2018.10.036. Epub 2018 Oct 12. Gene. 2019. PMID: 30321661
-
Post-translational regulation of the maternal-to-zygotic transition.Cell Mol Life Sci. 2018 May;75(10):1707-1722. doi: 10.1007/s00018-018-2750-y. Epub 2018 Feb 9. Cell Mol Life Sci. 2018. PMID: 29427077 Free PMC article. Review.
-
The parental contributions to early plant embryogenesis and the concept of maternal-to-zygotic transition in plants.Curr Opin Plant Biol. 2022 Feb;65:102144. doi: 10.1016/j.pbi.2021.102144. Epub 2021 Nov 22. Curr Opin Plant Biol. 2022. PMID: 34823206 Review.
Cited by
-
Echs1-mediated histone crotonylation facilitates zygotic genome activation and expression of repetitive elements in early mammalian embryos.Nat Commun. 2025 Jul 1;16(1):5630. doi: 10.1038/s41467-025-60565-z. Nat Commun. 2025. PMID: 40593492 Free PMC article.
-
Capillary Electrophoresis-Mass Spectrometry for Top-Down Proteomics.Annu Rev Anal Chem (Palo Alto Calif). 2025 May;18(1):125-147. doi: 10.1146/annurev-anchem-071124-092242. Epub 2025 Jan 23. Annu Rev Anal Chem (Palo Alto Calif). 2025. PMID: 39847747 Review.
-
Binding of zebrafish lipovitellin and L1‑ORF2 increases the accessibility of L1‑ORF2 via interference with histone wrapping.Int J Mol Med. 2025 Jan;55(1):2. doi: 10.3892/ijmm.2024.5443. Epub 2024 Oct 25. Int J Mol Med. 2025. PMID: 39450563 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

