A bioinformatic analysis of T-cell epitope diversity in SARS-CoV-2 variants: association with COVID-19 clinical severity in the United States population
- PMID: 38784379
- PMCID: PMC11112498
- DOI: 10.3389/fimmu.2024.1357731
A bioinformatic analysis of T-cell epitope diversity in SARS-CoV-2 variants: association with COVID-19 clinical severity in the United States population
Abstract
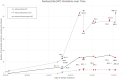
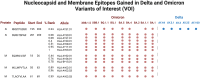
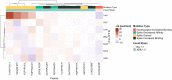
Long-term immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) requires the identification of T-cell epitopes affecting host immunogenicity. In this computational study, we explored the CD8+ epitope diversity estimated in 27 of the most common HLA-A and HLA-B alleles, representing most of the United States population. Analysis of 16 SARS-CoV-2 variants [B.1, Alpha (B.1.1.7), five Delta (AY.100, AY.25, AY.3, AY.3.1, AY.44), and nine Omicron (BA.1, BA.1.1, BA.2, BA.4, BA.5, BQ.1, BQ.1.1, XBB.1, XBB.1.5)] in analyzed MHC class I alleles revealed that SARS-CoV-2 CD8+ epitope conservation was estimated at 87.6%-96.5% in spike (S), 92.5%-99.6% in membrane (M), and 94.6%-99% in nucleocapsid (N). As the virus mutated, an increasing proportion of S epitopes experienced reduced predicted binding affinity: 70% of Omicron BQ.1-XBB.1.5 S epitopes experienced decreased predicted binding, as compared with ~3% and ~15% in the earlier strains Delta AY.100-AY.44 and Omicron BA.1-BA.5, respectively. Additionally, we identified several novel candidate HLA alleles that may be more susceptible to severe disease, notably HLA-A*32:01, HLA-A*26:01, and HLA-B*53:01, and relatively protected from disease, such as HLA-A*31:01, HLA-B*40:01, HLA-B*44:03, and HLA-B*57:01. Our findings support the hypothesis that viral genetic variation affecting CD8 T-cell epitope immunogenicity contributes to determining the clinical severity of acute COVID-19. Achieving long-term COVID-19 immunity will require an understanding of the relationship between T cells, SARS-CoV-2 variants, and host MHC class I genetics. This project is one of the first to explore the SARS-CoV-2 CD8+ epitope diversity that putatively impacts much of the United States population.
Keywords: CD8 T cell epitope; COVID-19; HLA; SARS-CoV-2; T cell epitope; bioinformatics; vaccine design.
Copyright © 2024 Kim, Elnaggar, Varnado, Feehan, Tauzier, Rose, Lamers, Sevalia, Nicholas, Gravois, Fort, Crabtree and Miele.
Conflict of interest statement
Authors RR and SL are employed by the company BioInfoExperts, LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

