First-principles study of molecular hydrogen binding to heme in competition with O2, NO and CO
- PMID: 38784410
- PMCID: PMC11110138
- DOI: 10.1039/d4ra02091j
First-principles study of molecular hydrogen binding to heme in competition with O2, NO and CO
Abstract
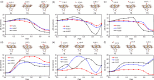
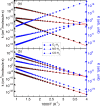
Molecular hydrogen shows antioxidant activity and distinct efficacy towards vascular diseases, but the understanding of this is not yet satisfactory at the atomic level. In this work, we study the binding properties of H2 to the heme group in relation with other diatomic molecules (DMs), including O2, NO and CO, and their displacement reactions, using first-principles calculations. We carry out molecular modeling of the heme group, using iron-porphyrin with the imidazole ligand, i.e., FePIm, and smaller models of Fe(CnHn+2N2)2NH3 with n = 3 and 1, and of molecular complexes of heme-DM and -H. Through analysis of optimized geometries and energetics, it is found that the order of binding strength of DMs or H to the Fe of heme is NO > O2 > CO > H > H2 for FePIm-based systems, while it is H > O2 > NO > CO > H2 for model-based systems. We calculate the activation energies for displacement reactions of H2 and H by other DMs, revealing that the H2 displacements occur spontaneously while the H displacements require a large amount of energy. Finally, our calculations corroborate that the rate constants increase with increasing temperature according to the Arrhenius relation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Structural basis for ligand discrimination and response initiation in the heme-based oxygen sensor FixL.Biochemistry. 1996 Jul 23;35(29):9539-48. doi: 10.1021/bi9530853. Biochemistry. 1996. PMID: 8755735
-
A density functional theory investigation of Fe-N-O bonding in heme proteins and model systems.J Am Chem Soc. 2003 Dec 31;125(52):16387-96. doi: 10.1021/ja030340v. J Am Chem Soc. 2003. PMID: 14692781
-
Binding of O2 and NO to heme in heme-nitric oxide/oxygen-binding (H-NOX) proteins. A theoretical study.J Phys Chem B. 2013 Sep 5;117(35):10103-14. doi: 10.1021/jp403998u. Epub 2013 Aug 22. J Phys Chem B. 2013. PMID: 23926882 Free PMC article.
-
VTST/MT studies of the catalytic mechanism of C-H activation by transition metal complexes with [Cu2(μ-O2)], [Fe2(μ-O2)] and Fe(IV)-O cores based on DFT potential energy surfaces.J Biol Inorg Chem. 2017 Apr;22(2-3):321-338. doi: 10.1007/s00775-017-1441-8. Epub 2017 Jan 16. J Biol Inorg Chem. 2017. PMID: 28091753 Review.
-
Photochemical reactions of cytochrome oxidase at low temperatures.Adv Biophys. 1978;11:285-308. Adv Biophys. 1978. PMID: 209671 Review.
Cited by
-
Revealing the flame retardancy of cotton fabrics treated with ammonium ethylenediamine tetramethylenephosphonate and trimethylol melamine.RSC Adv. 2025 Apr 4;15(14):10522-10531. doi: 10.1039/d5ra00402k. eCollection 2025 Apr 4. RSC Adv. 2025. PMID: 40190637 Free PMC article.
References
LinkOut - more resources
Full Text Sources

