Pyrazolopyridine-based kinase inhibitors for anti-cancer targeted therapy
- PMID: 38784451
- PMCID: PMC11110789
- DOI: 10.1039/d4md00003j
Pyrazolopyridine-based kinase inhibitors for anti-cancer targeted therapy
Abstract
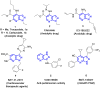
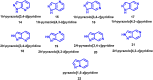
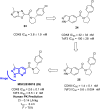
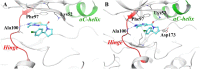
The need for effective cancer treatments continues to be a challenge for the biomedical research community. In this case, the advent of targeted therapy has significantly improved therapeutic outcomes. Drug discovery and development efforts targeting kinases have resulted in the approval of several small-molecule anti-cancer drugs based on ATP-mimicking heterocyclic cores. Pyrazolopyridines are a group of privileged heterocyclic cores in kinase drug discovery, which are present in several inhibitors that have been developed against various cancers. Notably, selpercatinib, glumetinib, camonsertib and olverembatinib have either received approval or are in late-phase clinical studies. This review presents the success stories employing pyrazolopyridine scaffolds as hinge-binding cores to address various challenges in kinase-targeted drug discovery research.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

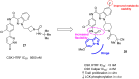
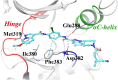
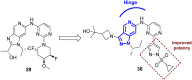

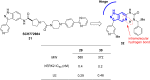
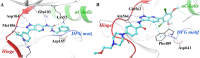
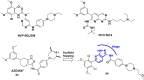
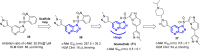
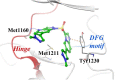
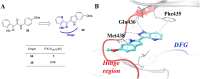
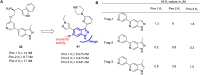

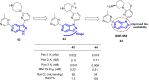
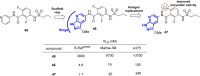
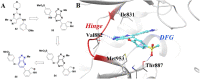
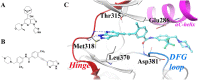
References
Publication types
LinkOut - more resources
Full Text Sources

