Imidazopyrimidine: from a relatively exotic scaffold to an evolving structural motif in drug discovery
- PMID: 38784469
- PMCID: PMC11110759
- DOI: 10.1039/d3md00718a
Imidazopyrimidine: from a relatively exotic scaffold to an evolving structural motif in drug discovery
Abstract
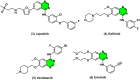
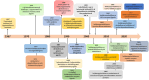
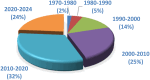
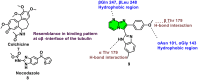
Nitrogen-fused heterocycles are of immense importance in modern drug discovery and development. Among them, imidazopyrimidine is a highly versatile scaffold with vast pharmacological utility. These compounds demonstrate a broad spectrum of pharmacological actions, including antiviral, antifungal, anti-inflammatory, and anticancer. Their adaptable structure allows for extensive structural modifications, which can be utilized for optimizing pharmacological effects via structure-activity relationship (SAR) studies. Additionally, imidazopyrimidine derivatives are particularly noteworthy for their ability to target specific molecular entities, such as protein kinases, which are crucial components of various cellular signaling pathways associated with multiple diseases. Despite the evident importance of imidazopyrimidines in drug discovery, there is a notable lack of a comprehensive review that outlines their role in this field. This review highlights the ongoing interest and investment in exploring the therapeutic potential of imidazopyrimidine compounds, underscoring their pivotal role in shaping the future of drug discovery and clinical medicine.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures










References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

