Brain organoids engineered to give rise to glia and neural networks after 90 days in culture exhibit human-specific proteoforms
- PMID: 38784709
- PMCID: PMC11111902
- DOI: 10.3389/fncel.2024.1383688
Brain organoids engineered to give rise to glia and neural networks after 90 days in culture exhibit human-specific proteoforms
Abstract
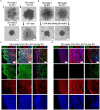
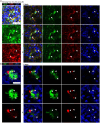
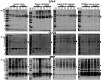
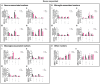
Human brain organoids are emerging as translationally relevant models for the study of human brain health and disease. However, it remains to be shown whether human-specific protein processing is conserved in human brain organoids. Herein, we demonstrate that cell fate and composition of unguided brain organoids are dictated by culture conditions during embryoid body formation, and that culture conditions at this stage can be optimized to result in the presence of glia-associated proteins and neural network activity as early as three-months in vitro. Under these optimized conditions, unguided brain organoids generated from induced pluripotent stem cells (iPSCs) derived from male-female siblings are similar in growth rate, size, and total protein content, and exhibit minimal batch-to-batch variability in cell composition and metabolism. A comparison of neuronal, microglial, and macroglial (astrocyte and oligodendrocyte) markers reveals that profiles in these brain organoids are more similar to autopsied human cortical and cerebellar profiles than to those in mouse cortical samples, providing the first demonstration that human-specific protein processing is largely conserved in unguided brain organoids. Thus, our organoid protocol provides four major cell types that appear to process proteins in a manner very similar to the human brain, and they do so in half the time required by other protocols. This unique copy of the human brain and basic characteristics lay the foundation for future studies aiming to investigate human brain-specific protein patterning (e.g., isoforms, splice variants) as well as modulate glial and neuronal processes in an in situ-like environment.
Keywords: astrocyte; heterogeneity; human brain; microglia; mouse brain; oligodendrocytes; protein processing; species differences.
Copyright © 2024 Wenzel and Mousseau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Dynamic Characterization of Structural, Molecular, and Electrophysiological Phenotypes of Human-Induced Pluripotent Stem Cell-Derived Cerebral Organoids, and Comparison with Fetal and Adult Gene Profiles.Cells. 2020 May 23;9(5):1301. doi: 10.3390/cells9051301. Cells. 2020. PMID: 32456176 Free PMC article.
-
A Robust Pipeline for the Multi-Stage Accelerated Differentiation of Functional 3D Cortical Organoids from Human Pluripotent Stem Cells.Curr Protoc. 2023 Jan;3(1):e641. doi: 10.1002/cpz1.641. Curr Protoc. 2023. PMID: 36633423 Free PMC article.
-
Dynamic 3D Combinatorial Generation of hPSC-Derived Neuromesodermal Organoids With Diverse Regional and Cellular Identities.Curr Protoc. 2022 Oct;2(10):e568. doi: 10.1002/cpz1.568. Curr Protoc. 2022. PMID: 36264199 Free PMC article.
-
Human neural organoids: Models for developmental neurobiology and disease.Dev Biol. 2021 Oct;478:102-121. doi: 10.1016/j.ydbio.2021.06.012. Epub 2021 Jun 25. Dev Biol. 2021. PMID: 34181916 Free PMC article. Review.
-
Application of Human Brain Organoids-Opportunities and Challenges in Modeling Human Brain Development and Neurodevelopmental Diseases.Int J Mol Sci. 2023 Aug 7;24(15):12528. doi: 10.3390/ijms241512528. Int J Mol Sci. 2023. PMID: 37569905 Free PMC article. Review.
Cited by
-
Induced Microglial-like Cells Derived from Familial and Sporadic Alzheimer's Disease Peripheral Blood Monocytes Show Abnormal Phagocytosis and Inflammatory Response to PSEN1 E280A Cholinergic-like Neurons.Int J Mol Sci. 2025 Jul 24;26(15):7162. doi: 10.3390/ijms26157162. Int J Mol Sci. 2025. PMID: 40806295 Free PMC article.
-
Human brain organoids containing microglia that have arisen innately adapt to a β-amyloid challenge better than those in which microglia are integrated by co-culture.Stem Cell Res Ther. 2024 Aug 13;15(1):258. doi: 10.1186/s13287-024-03876-0. Stem Cell Res Ther. 2024. PMID: 39135132 Free PMC article.
References
-
- Bayés À., Collins M. O., Croning M. D. R., van de Lagemaat L. N., Choudhary J. S., Grant S. G. N. (2012). Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One 7:e46683. doi: 10.1371/journal.pone.0046683 - DOI - PMC - PubMed
-
- Bellinger F. P., Bellinger M. T., Seale L. A., Takemoto A. S., Raman A. V., Miki T., et al. . (2011). Glutathione peroxidase 4 is associated with neuromelanin in substantia nigra and dystrophic axons in putamen of Parkinson’s brain. Mol. Neurodegener. 6:8. doi: 10.1186/1750-1326-6-8 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

