The distribution and antibiotic-resistant characteristics and risk factors of pathogens associated with clinical biliary tract infection in humans
- PMID: 38784792
- PMCID: PMC11112516
- DOI: 10.3389/fmicb.2024.1404366
The distribution and antibiotic-resistant characteristics and risk factors of pathogens associated with clinical biliary tract infection in humans
Abstract
Introduction: Biliary Infection in patients is a common and important phenomenon resulting in severe complications and high morbidity, while the distributions and drug resistance profiles of biliary bacteria and related risk factors are dynamic. This study explored the characteristics of and risk factors for biliary infection to promote the rational use of antibiotics in clinically.
Methods: Bacterial identification and drug susceptibility testing were completed using the Vitek 2 Compact analysis system. The distribution and antibiotic-resistant characteristics of 3,490 strains of biliary bacteria in patients at Nankai Hospital from 2019 to 2021 were analyzed using Whonet 5.6 and SPSS 26.0 software. We then retrospectively analyzed the clinical data and risk factors associated with 2,340 strains of Gram-negative bacilli, which were divided into multidrug-resistant bacteria (1,508 cases) and non-multidrug-resistant bacteria (832 cases) by a multivariate Cox regression model.
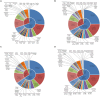
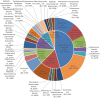
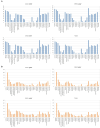
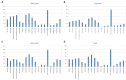
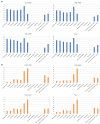
Results and discussion: A total of 3,490 pathogenic bacterial strains were isolated from bile samples, including 2,340 (67.05%) Gram-negative strains, 1,029 (29.48%) Gram-positive strains, and 109 (4.56%) fungal strains. The top five pathogenic bacteria were Escherichia coli, Klebsiella pneumoniae, Enterococcus faecium, Enterococcus faecalis, and Pseudomonas aeruginosa. The rate of Escherichia coli resistance to ciprofloxacin increased (p < 0.05), while the resistance to amikacin decreased (p < 0.05). The resistance of Klebsiella pneumoniae to cephalosporins, carbapenems, β-lactamase inhibitors, cephalases, aminoglycosides, and quinolones increased (p < 0.05), and the resistance of Pseudomonas aeruginosa to piperacillin, piperacillin/tazobactam, ticacillin/clavulanic acid, and amicacin declined significantly (p < 0.05). The resistance of Enterococcus faecium to tetracycline increased by year (p < 0.05), and the resistance of Enterococcus faecalis to erythromycin and high-concentration gentamicin declined (p < 0.05). Multivariate logistic regression analysis suggested that the administration of third- or fourth-generation cephalosporins was an independent risk factor for biliary infection. In summary, Gram-negative bacilli were the most common pathogenic bacteria isolated from biliary infection patients, especially Escherichia coli, and the rates and patterns of drug resistance were high and in constant flux; therefore, rational antimicrobial drug use should be carried out considering risk factors.
Keywords: Gram-negative bacteria; biliary pathogens; distribution of bacteria; multidrug-resistant bacteria; risk factors.
Copyright © 2024 Chen, Lai, Song, Lu, Liang, Ouyang, Zheng, Chen, Yin, Li and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Alreshidi M., Dunstan H., Roberts T., Alreshidi F., Hossain A., Bardakci F., et al. . (2023). Cytoplasmic amino acid profiles of clinical and ATCC 29213 strains of Staphylococcus aureus harvested at different growth phases. Biomol. Biomed. 23, 1038–1050. doi: 10.17305/bb.2023.9246 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

