Diversity, evolution, and emergence of fish viruses
- PMID: 38785422
- PMCID: PMC11237817
- DOI: 10.1128/jvi.00118-24
Diversity, evolution, and emergence of fish viruses
Abstract
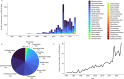
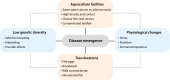
The production of aquatic animals has more than doubled over the last 50 years and is anticipated to continually increase. While fish are recognized as a valuable and sustainable source of nutrition, particularly in the context of human population growth and climate change, the rapid expansion of aquaculture coincides with the emergence of highly pathogenic viruses that often spread globally through aquacultural practices. Here, we provide an overview of the fish virome and its relevance for disease emergence, with a focus on the insights gained through metagenomic sequencing, noting potential areas for future study. In particular, we describe the diversity and evolution of fish viruses, for which the majority have no known disease associations, and demonstrate how viruses emerge in fish populations, most notably at an expanding domestic-wild interface. We also show how wild fish are a powerful and tractable model system to study virus ecology and evolution more broadly and can be used to identify the major factors that shape vertebrate viromes. Central to this is a process of virus-host co-divergence that proceeds over many millions of years, combined with ongoing cross-species virus transmission.
Keywords: aquaculture; ecology; emerging disease; evolution; fish; virome; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Complex dynamics at the interface between wild and domestic viruses of finfish.Curr Opin Virol. 2011 Jul;1(1):73-80. doi: 10.1016/j.coviro.2011.05.010. Epub 2011 Jun 17. Curr Opin Virol. 2011. PMID: 22440571 Review.
-
Five Species of Wild Freshwater Sport Fish in Wisconsin, USA, Reveal Highly Diverse Viromes.Pathogens. 2024 Feb 7;13(2):150. doi: 10.3390/pathogens13020150. Pathogens. 2024. PMID: 38392888 Free PMC article.
-
Novel viruses discovered in metatranscriptomic analysis of farmed barramundi in Asia and Australia.Virology. 2024 Nov;599:110208. doi: 10.1016/j.virol.2024.110208. Epub 2024 Aug 14. Virology. 2024. PMID: 39154629
-
Emerging viruses in aquaculture.Curr Opin Virol. 2019 Feb;34:97-103. doi: 10.1016/j.coviro.2018.12.008. Epub 2019 Feb 1. Curr Opin Virol. 2019. PMID: 30711892 Review.
-
Virome composition in marine fish revealed by meta-transcriptomics.Virus Evol. 2021 Feb 4;7(1):veab005. doi: 10.1093/ve/veab005. eCollection 2021 Jan. Virus Evol. 2021. PMID: 33623709 Free PMC article.
Cited by
-
RNA-Sequencing Analysis of the Viral Community in Yellow Catfish (Pelteobagrus fulvidraco) in the Upper Reaches of the Yangtze River.Animals (Basel). 2024 Nov 25;14(23):3386. doi: 10.3390/ani14233386. Animals (Basel). 2024. PMID: 39682352 Free PMC article.
-
Multiple Horizontal Transfers of Immune Genes Between Distantly Related Teleost Fishes.Mol Biol Evol. 2025 Apr 30;42(5):msaf107. doi: 10.1093/molbev/msaf107. Mol Biol Evol. 2025. PMID: 40378191 Free PMC article.
-
Viral Threats to Australian Fish and Prawns: Economic Impacts and Biosecurity Solutions-A Systematic Review.Viruses. 2025 May 10;17(5):692. doi: 10.3390/v17050692. Viruses. 2025. PMID: 40431703 Free PMC article. Review.
References
-
- Miller AK, Mifsud JCO, Costa VA, Grimwood RM, Kitson J, Baker C, Brosnahan CL, Pande A, Holmes EC, Gemmell NJ, Geoghegan JL. 2021. Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the Coronaviridae. Virus Evol 7:veab050. doi: 10.1093/ve/veab050 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

