A novel multiplex fluorescent-labeling method for the visualization of mixed-species biofilms in vitro
- PMID: 38785429
- PMCID: PMC11218471
- DOI: 10.1128/spectrum.00253-24
A novel multiplex fluorescent-labeling method for the visualization of mixed-species biofilms in vitro
Abstract
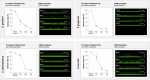
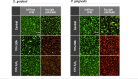
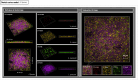
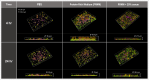
In nature, bacteria usually exist as mixed-species biofilms, where they engage in a range of synergistic and antagonistic interactions that increase their resistance to environmental challenges. Biofilms are a major cause of persistent infections, and dispersal from initial foci can cause new infections at distal sites thus warranting further investigation. Studies of development and spatial interactions in mixed-species biofilms can be challenging due to difficulties in identifying the different bacterial species in situ. Here, we apply CellTrace dyes to studies of biofilm bacteria and present a novel application for multiplex labeling, allowing identification of different bacteria in mixed-species, in vitro biofilm models. Oral bacteria labeled with CellTrace dyes (far red, yellow, violet, and CFSE [green]) were used to create single- and mixed-species biofilms, which were analyzed with confocal spinning disk microscopy (CSDM). Biofilm supernatants were studied with flow cytometry (FC). Both Gram-positive and Gram-negative bacteria were well labeled and CSDM revealed biofilms with clear morphology and stable staining for up to 4 days. Analysis of CellTrace labeled cells in supernatants using FC showed differences in the biofilm dispersal between bacterial species. Multiplexing with different colored dyes allowed visualization of spatial relationships between bacteria in mixed-species biofilms and relative coverage by the different species was revealed through segmentation of the CSDM images. This novel application, thus, offers a powerful tool for studying structure and composition of mixed-species biofilms in vitro.IMPORTANCEAlthough most chronic infections are caused by mixed-species biofilms, much of our knowledge still comes from planktonic cultures of single bacterial species. Studies of formation and development of mixed-species biofilms are, therefore, required. This work describes a method applicable to labeling of bacteria for in vitro studies of biofilm structure and dispersal. Critically, labeled bacteria can be multiplexed for identification of different species in mixed-species biofilms using confocal spinning disk microscopy, facilitating investigation of biofilm development and spatial interactions under different environmental conditions. The study is an important step in increasing the tools available for such complex and challenging studies.
Keywords: CellTrace; biofilm detachment; biofilm growth; confocal spinning disc microscopy; flow cytometry; live imaging; microscopy; oral bacteria; oral disease; staining.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Multiplex FISH analysis of a six-species bacterial biofilm.J Microbiol Methods. 2004 Jan;56(1):37-47. doi: 10.1016/j.mimet.2003.09.003. J Microbiol Methods. 2004. PMID: 14706749
-
The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods.J Diabetes Res. 2014;2014:153586. doi: 10.1155/2014/153586. Epub 2014 Apr 15. J Diabetes Res. 2014. PMID: 24839608 Free PMC article.
-
Biofilm growth and IL-8 & TNF-α-inducing properties of Candida albicans in the presence of oral gram-positive and gram-negative bacteria.BMC Microbiol. 2020 Jun 11;20(1):156. doi: 10.1186/s12866-020-01834-3. BMC Microbiol. 2020. PMID: 32527216 Free PMC article.
-
Biofilm patterns in gram-positive and gram-negative bacteria.Microbiol Res. 2021 Oct;251:126829. doi: 10.1016/j.micres.2021.126829. Epub 2021 Jul 23. Microbiol Res. 2021. PMID: 34332222 Review.
-
Biofilm formation as microbial development.Annu Rev Microbiol. 2000;54:49-79. doi: 10.1146/annurev.micro.54.1.49. Annu Rev Microbiol. 2000. PMID: 11018124 Review.
Cited by
-
MUC5B modulation of early oral biofilm glucose metabolism.Front Oral Health. 2025 Feb 11;6:1516025. doi: 10.3389/froh.2025.1516025. eCollection 2025. Front Oral Health. 2025. PMID: 40008185 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

