Surveillance and phylogenetic analysis of a pathogenic bacterium candidate in nasal discharge from children
- PMID: 38785433
- PMCID: PMC11218501
- DOI: 10.1128/spectrum.00566-24
Surveillance and phylogenetic analysis of a pathogenic bacterium candidate in nasal discharge from children
Abstract
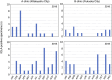
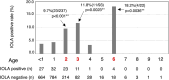
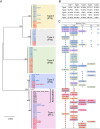
"The infectious organism lurking in human airways (IOLA)" is a candidate pathogenic bacterium detected in bronchoalveolar lavage fluid specimens derived from adult patients with chronic lower respiratory tract infections. Genomic analyses of IOLA have revealed that it possesses the smallest and most AT-rich genome among human-derived bacteria. However, its biological properties remain unclear because no culture method has been established for IOLA. Here, we conducted a large-scale IOLA surveillance study of nasal discharge specimens from children in Japan and investigated the correlation between IOLA detection frequency and patient characteristics. We detected IOLA in 5.4% (103 of 1,920) of pediatric nasal discharge samples. No significant differences were observed in the frequency of detection based on the patient's background. However, with respect to age, the frequency of detection tended to be significantly high at 2-3 and 6 years old. Phylogenetic analysis revealed five phylotypes in the IOLA 16S rRNA gene sequences, and the sequences detected in adult patients with respiratory infections in a previous study belonged to one of the five phylotypes. The involvement of IOLA in the symptoms is not clear, but IOLA is detected at a relatively high frequency in pediatric nasal discharge. Many subjects with detected IOLA were not always IOLA positive, and IOLA was detected transiently. Our findings suggest that IOLA is horizontally transmitted through groups in nursery and elementary schools, and there are differences in biological characteristics among the IOLA phylotypes.IMPORTANCE"The infectious organism lurking in human airways (IOLA)" is a candidate pathogenic bacterium strongly suspected to be infectious to the respiratory tracts of humans and animals. However, a culture method for IOLA has not been established yet, and its properties remain unclear. In this study, IOLA was detected at a relatively high frequency in the nasal discharge of children, and five phylotypes of IOLA were identified. One of these phylotypes was found in the bronchoalveolar lavage fluid from adult patients, suggesting lineage-specific differences in the pathogenicity of IOLA. Moreover, it was suggested that IOLA is horizontally transmitted when children gather in groups such as nursery and elementary schools. These findings strongly indicate that IOLAs have been clinically undetected so far but are spreading among children, with one lineage being involved in respiratory diseases in adults. Examining the presence of IOLA in clinical specimens may help to understand the etiology of respiratory diseases with unknown causes.
Keywords: 16S rRNA gene; infectious disease; uncultured bacterium.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
An unclassified microorganism: novel pathogen candidate lurking in human airways.PLoS One. 2014 Jul 31;9(7):e103646. doi: 10.1371/journal.pone.0103646. eCollection 2014. PLoS One. 2014. PMID: 25080337 Free PMC article.
-
A human respiratory tract-associated bacterium with an extremely small genome.Commun Biol. 2021 May 26;4(1):628. doi: 10.1038/s42003-021-02162-6. Commun Biol. 2021. PMID: 34040152 Free PMC article.
-
Three clinically distinct chronic pediatric airway infections share a common core microbiota.Ann Am Thorac Soc. 2014 Sep;11(7):1039-48. doi: 10.1513/AnnalsATS.201312-456OC. Ann Am Thorac Soc. 2014. PMID: 24597615 Free PMC article.
-
Perspective on the clone library method for infectious diseases.Respir Investig. 2021 Nov;59(6):741-747. doi: 10.1016/j.resinv.2021.07.003. Epub 2021 Aug 14. Respir Investig. 2021. PMID: 34400128 Review.
-
Bacterial microbiota of the nasal passages across the span of human life.Curr Opin Microbiol. 2018 Feb;41:8-14. doi: 10.1016/j.mib.2017.10.023. Epub 2017 Nov 20. Curr Opin Microbiol. 2018. PMID: 29156371 Free PMC article. Review.
References
-
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi:10.1038/nature12352 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

