Highly sensitive extraction-free saliva-based molecular assay for rapid diagnosis of SARS-CoV-2
- PMID: 38785448
- PMCID: PMC11237525
- DOI: 10.1128/jcm.00600-24
Highly sensitive extraction-free saliva-based molecular assay for rapid diagnosis of SARS-CoV-2
Abstract
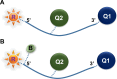
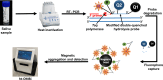
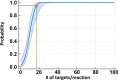
The COVID-19 pandemic highlighted the necessity of fast, sensitive, and efficient methods to test large populations for respiratory viruses. The "gold standard" molecular assays for detecting respiratory viruses, such as quantitative polymerase chain reaction (qPCR) and reverse transcription qPCR (RT-qPCR), rely on invasive swab samples and require time-consuming and labor-intensive extraction processes. Moreover, the turnaround time for RT-qPCR-based assays is too lengthy for rapid screening. Extraction-free saliva-based methods provide a non-invasive sampling process with a fast turnaround time and are suitable for high-throughput applications. However, when used with a standard RT-qPCR system, the absence of extraction significantly reduces the assays' sensitivity. Here, using a novel optical modulation biosensing (OMB) platform, we developed a rapid and highly sensitive extraction-free saliva-based molecular assay. We blindly tested 364 paired nasopharyngeal swabs and saliva samples from suspected SARS-CoV-2 cases in Israel. Compared with the gold standard swab-based RT-qPCR assay, the sensitivity of the extraction-free saliva-based OMB assay is 90.7%, much higher than the sensitivity of extraction-free saliva-based RT-qPCR assay (77.8%) with similar specificity (95.3% and 97.6%, respectively). Moreover, out of 12 samples identified by the OMB-based assay as positive, 8 samples were collected from hospitalized patients in a COVID-19 ward and were verified to be SARS-CoV-2-positive upon admission, indicating that the actual clinical sensitivity and specificity of the OMB assay are higher. Considering its user-friendly saliva-based protocol, short and cost-effective extraction-free process, and high clinical accuracy, the OMB-based molecular assay is very suitable for high-throughput testing of large populations for respiratory viruses.
Importance: Three years after the SARS-CoV-2 outbreak, there are no molecular tests that combine low-cost and straightforward sample preparation, effective sample handling, minimal reagent and disposable requirements, high sensitivity, and high throughput required for mass screening. Existing rapid molecular techniques typically sacrifice certain requirements to meet others. Yet, localized outbreaks of novel viral diseases happen daily in different parts of the world. In this context, respiratory diseases are of specific importance, as they are frequently airborne and highly contagious, with the potential for a rapid global spread. The widely accepted opinion is that another pandemic is just a question of time. To ensure that the containment efforts for the upcoming "disease X" are successful, introducing rapid, high-throughput, and highly sensitive diagnostic methods for detecting and identifying pathogens is critical. A few months into the pandemic, saliva was suggested as a diagnostic matrix for SARS-CoV-2 detection. The collection of saliva does not require swabs and is minimally invasive. In particular, extraction-free saliva-based assays require fewer reagents and disposables, and therefore are faster and cheaper, offering an appealing alternative for low-income countries. Unfortunately, current extraction-free saliva-based detection methods, such as direct RT-qPCR or isothermal amplification, have either low sensitivity or low throughput. Therefore, we believe that the presented highly sensitive ht-OMBi platform and the extraction-free saliva-based molecular assay can become an essential tool in the infectious disease monitoring toolbox.
Keywords: COVID-19; MMB; SARS-CoV-2; assay; detection; diagnostics; extraction-free; high-throughput; highly sensitive; ht-OMBi; optical modulation biosensing; saliva-based.
Conflict of interest statement
A.D. has a financial interest in MagBiosense, Inc. which, however, did not provide funding for this work
Figures



Similar articles
-
Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa.Lancet Glob Health. 2025 Jul;13(7):e1258-e1267. doi: 10.1016/S2214-109X(25)00150-0. Lancet Glob Health. 2025. PMID: 40580991 Free PMC article.
-
Multisite Clinical Validation of Isothermal Amplification-Based SARS-CoV-2 Detection Assays Using Different Sampling Strategies.Microbiol Spectr. 2021 Oct 31;9(2):e0084621. doi: 10.1128/Spectrum.00846-21. Epub 2021 Oct 20. Microbiol Spectr. 2021. PMID: 34668736 Free PMC article.
-
Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification.Clin Chem. 2021 Jan 30;67(2):415-424. doi: 10.1093/clinchem/hvaa267. Clin Chem. 2021. PMID: 33098427 Free PMC article.
-
Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis.PLoS One. 2021 Jun 10;16(6):e0253007. doi: 10.1371/journal.pone.0253007. eCollection 2021. PLoS One. 2021. PMID: 34111196 Free PMC article.
-
Diagnostic performance of different sampling approaches for SARS-CoV-2 RT-PCR testing: a systematic review and meta-analysis.Lancet Infect Dis. 2021 Sep;21(9):1233-1245. doi: 10.1016/S1473-3099(21)00146-8. Epub 2021 Apr 12. Lancet Infect Dis. 2021. PMID: 33857405 Free PMC article.
Cited by
-
Rapid and Highly Sensitive Detection of Ricin in Biological Fluids Using Optical Modulation Biosensing.Biosensors (Basel). 2025 May 6;15(5):295. doi: 10.3390/bios15050295. Biosensors (Basel). 2025. PMID: 40422034 Free PMC article.
-
Sensitivity and Cross-Reactivity Analysis of Serotype-Specific Anti-NS1 Serological Assays for Dengue Virus Using Optical Modulation Biosensing.Biosensors (Basel). 2025 Jul 14;15(7):453. doi: 10.3390/bios15070453. Biosensors (Basel). 2025. PMID: 40710103 Free PMC article.
References
-
- Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing. 2021. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specim.... Retrieved 22 May 2022.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

