Stochastic Compartment Model with Mortality and Its Application to Epidemic Spreading in Complex Networks
- PMID: 38785610
- PMCID: PMC11120256
- DOI: 10.3390/e26050362
Stochastic Compartment Model with Mortality and Its Application to Epidemic Spreading in Complex Networks
Abstract
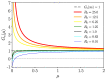
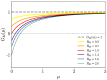
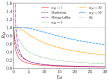
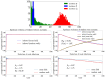
We study epidemic spreading in complex networks by a multiple random walker approach. Each walker performs an independent simple Markovian random walk on a complex undirected (ergodic) random graph where we focus on the Barabási-Albert (BA), Erdös-Rényi (ER), and Watts-Strogatz (WS) types. Both walkers and nodes can be either susceptible (S) or infected and infectious (I), representing their state of health. Susceptible nodes may be infected by visits of infected walkers, and susceptible walkers may be infected by visiting infected nodes. No direct transmission of the disease among walkers (or among nodes) is possible. This model mimics a large class of diseases such as Dengue and Malaria with the transmission of the disease via vectors (mosquitoes). Infected walkers may die during the time span of their infection, introducing an additional compartment D of dead walkers. Contrary to the walkers, there is no mortality of infected nodes. Infected nodes always recover from their infection after a random finite time span. This assumption is based on the observation that infectious vectors (mosquitoes) are not ill and do not die from the infection. The infectious time spans of nodes and walkers, and the survival times of infected walkers, are represented by independent random variables. We derive stochastic evolution equations for the mean-field compartmental populations with the mortality of walkers and delayed transitions among the compartments. From linear stability analysis, we derive the basic reproduction numbers RM,R0 with and without mortality, respectively, and prove that RM<R0. For RM,R0>1, the healthy state is unstable, whereas for zero mortality, a stable endemic equilibrium exists (independent of the initial conditions), which we obtained explicitly. We observed that the solutions of the random walk simulations in the considered networks agree well with the mean-field solutions for strongly connected graph topologies, whereas less well for weakly connected structures and for diseases with high mortality. Our model has applications beyond epidemic dynamics, for instance in the kinetics of chemical reactions, the propagation of contaminants, wood fires, and others.
Keywords: compartment model with mortality; epidemic spreading; memory effects; random graphs; random walks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
A Markovian random walk model of epidemic spreading.Contin Mech Thermodyn. 2021;33(4):1207-1221. doi: 10.1007/s00161-021-00970-z. Epub 2021 Jan 16. Contin Mech Thermodyn. 2021. PMID: 34776647 Free PMC article.
-
Random walks with stochastic resetting in complex networks: A discrete-time approach.Chaos. 2025 Jan 1;35(1):013119. doi: 10.1063/5.0238517. Chaos. 2025. PMID: 39787289
-
Four-compartment epidemic model with retarded transition rates.Phys Rev E. 2023 Apr;107(4-1):044207. doi: 10.1103/PhysRevE.107.044207. Phys Rev E. 2023. PMID: 37198844
-
Epidemics of random walkers in metapopulation model for complete, cycle, and star graphs.J Theor Biol. 2018 Aug 7;450:66-75. doi: 10.1016/j.jtbi.2018.04.029. Epub 2018 Apr 25. J Theor Biol. 2018. PMID: 29702109
-
Random walks with long-range memory on networks.Chaos. 2025 Jan 1;35(1):013117. doi: 10.1063/5.0243892. Chaos. 2025. PMID: 39787287
References
-
- Rhodes P., Bryant J.H. Public Health. Encyclopedia Britannica. 2024. [(accessed on 22 April 2024)]. Available online: https://www.britannica.com/topic/public-health.
-
- Kermack W.O., McKendrick A.G. A contribution to the mathematical theory of epidemics. Proc. Roy. Soc. A. 1927;115:700–721.
-
- Anderson R.M., May R.M. Infectious Diseases in Humans. Oxford University Press; Oxford, UK: 1992.
LinkOut - more resources
Full Text Sources

