Early Animal Origin of BACE1 APP/Aβ Proteolytic Function
- PMID: 38785802
- PMCID: PMC11117577
- DOI: 10.3390/biology13050320
Early Animal Origin of BACE1 APP/Aβ Proteolytic Function
Abstract
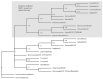
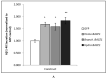
Alzheimer's disease is characterized, in part, by the accumulation of β-amyloid (Aβ) in the brain. Aβ is produced via the proteolysis of APP by BACE1 and γ-secretase. Since BACE1 is the rate-limiting enzyme in the production of Aβ, and a target for therapeutics, it is of interest to know when its proteolytic function evolved and for what purpose. Here, we take a functional evolutionary approach to show that BACE1 likely evolved from a gene duplication event near the base of the animal clade and that BACE1 APP/Aβ proteolytic function evolved during early animal diversification, hundreds of millions of years before the evolution of the APP/Aβ substrate. Our examination of BACE1 APP/Aβ proteolytic function includes cnidarians, ctenophores, and choanoflagellates. The most basal BACE1 ortholog is found in cnidarians, while ctenophores, placozoa, and choanoflagellates have genes equally orthologous to BACE1 and BACE2. BACE1 from a cnidarian (Hydra) can cleave APP to release Aβ, pushing back the date of the origin of its function to near the origin of animals. We tested more divergent BACE1/2 genes from a ctenophore (Mnemiopsis) and a choanoflagellate (Monosiga), and neither has this activity. These findings indicate that the specific proteolytic function of BACE1 evolved during the very earliest diversification of animals, most likely after a gene-duplication event.
Keywords: BACE; amyloid precursor protein (APP); animal; evolution; β-amyloid (Aβ).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Vassar R., Bennett B.D., Babu-Khan S., Kahn S., Mendiaz E.A., Denis P., Teplow D.B., Ross S., Amarante P., Loeloff R., et al. Beta-Secretase Cleavage of Alzheimer’s Amyloid Precursor Protein by the Transmembrane Aspartic Protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

