Herpesvirus Entry Mediator as an Immune Checkpoint Target and a Potential Prognostic Biomarker in Myeloid and Lymphoid Leukemia
- PMID: 38785930
- PMCID: PMC11117912
- DOI: 10.3390/biom14050523
Herpesvirus Entry Mediator as an Immune Checkpoint Target and a Potential Prognostic Biomarker in Myeloid and Lymphoid Leukemia
Abstract
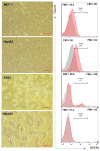
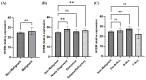
Herpesvirus entry mediator (HVEM) is a molecular switch that can modulate immune responses against cancer. The significance of HVEM as an immune checkpoint target and a potential prognostic biomarker in malignancies is still controversial. This study aims to determine whether HVEM is an immune checkpoint target with inhibitory effects on anti-tumor CD4+ T cell responses in vitro and whether HVEM gene expression is dysregulated in patients with acute lymphocytic leukemia (ALL). HVEM gene expression in tumor cell lines and peripheral blood mononuclear cells (PBMCs) from ALL patients and healthy controls was measured using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Tumor cells were left untreated (control) or were treated with an HVEM blocker before co-culturing with CD4+ T cells in vitro in a carboxyfluorescein succinimidyl ester (CFSE)-dependent proliferation assay. HVEM expression was upregulated in the chronic myelogenous leukemia cell line (K562) (FC = 376.3, p = 0.086) compared with normal embryonic kidney cells (Hek293). CD4+ T cell proliferation was significantly increased in the HVEM blocker-treated K562 cells (p = 0.0033). Significant HVEM differences were detected in ALL PBMCs compared with the controls, and these were associated with newly diagnosed ALL (p = 0.0011) and relapsed/refractory (p = 0.0051) B cell ALL (p = 0.0039) patients. A significant differentiation between malignant ALL and the controls was observed in a receiver operating characteristic (ROC) curve analysis with AUC = 0.78 ± 0.092 (p = 0.014). These results indicate that HVEM is an inhibitory molecule that may serve as a target for immunotherapy and a potential ALL biomarker.
Keywords: CD4+ T cells; acute lymphocytic leukemia; co-inhibitory molecules; herpesvirus entry mediator; immune checkpoint blockade; immune checkpoint receptor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Significant involvement of herpesvirus entry mediator in human esophageal squamous cell carcinoma.Cancer. 2014 Mar 15;120(6):808-17. doi: 10.1002/cncr.28491. Epub 2013 Nov 18. Cancer. 2014. PMID: 24249528
-
HVEM in acute lymphocytic leukemia facilitates tumour immune escape by inhibiting CD8+ T cell function.Cell Oncol (Dordr). 2024 Oct;47(5):1779-1796. doi: 10.1007/s13402-024-00959-1. Epub 2024 May 29. Cell Oncol (Dordr). 2024. PMID: 38809326
-
RNA and protein expression of herpesvirus entry mediator (HVEM) is associated with molecular markers, immunity-related pathways and relapse-free survival of patients with AML.Cancer Immunol Immunother. 2015 Dec;64(12):1505-15. doi: 10.1007/s00262-015-1755-8. Epub 2015 Sep 16. Cancer Immunol Immunother. 2015. PMID: 26377688 Free PMC article.
-
Modulation of T cell proliferation through the LIGHT-HVEM-BTLA cosignaling pathway.Recent Pat DNA Gene Seq. 2009;3(3):177-82. doi: 10.2174/187221509789318342. Recent Pat DNA Gene Seq. 2009. PMID: 19702559 Review.
-
The role of the BTLA-HVEM complex in the pathogenesis of breast cancer.Breast Cancer. 2024 May;31(3):358-370. doi: 10.1007/s12282-024-01557-7. Epub 2024 Mar 14. Breast Cancer. 2024. PMID: 38483699 Review.
References
-
- Sordo-Bahamonde C., Lorenzo-Herrero S., Granda-Díaz R., Martínez-Pérez A., Aguilar-García C., Rodrigo J.P., García-Pedrero J.M., Gonzalez S. Beyond the anti-PD-1/PD-L1 era: Promising role of the BTLA/HVEM axis as a future target for cancer immunotherapy. Mol. Cancer. 2023;22:142. doi: 10.1186/s12943-023-01845-4. - DOI - PMC - PubMed
-
- Sharma P., Siddiqui B.A., Anandhan S., Yadav S.S., Subudhi S.K., Gao J., Goswami S., Allison J.P. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021;11:838–857. doi: 10.1158/2159-8290.CD-20-1680. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

