Unraveling the Etiology of Dilated Cardiomyopathy through Differential miRNA-mRNA Interactome
- PMID: 38785931
- PMCID: PMC11117812
- DOI: 10.3390/biom14050524
Unraveling the Etiology of Dilated Cardiomyopathy through Differential miRNA-mRNA Interactome
Abstract
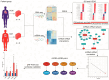
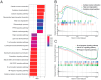
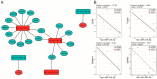
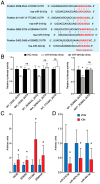
Dilated cardiomyopathy (DCM) encompasses various acquired or genetic diseases sharing a common phenotype. The understanding of pathogenetic mechanisms and the determination of the functional effects of each etiology may allow for tailoring different therapeutic strategies. MicroRNAs (miRNAs) have emerged as key regulators in cardiovascular diseases, including DCM. However, their specific roles in different DCM etiologies remain elusive. Here, we applied mRNA-seq and miRNA-seq to identify the gene and miRNA signature from myocardial biopsies from four patients with DCM caused by volume overload (VCM) and four with ischemic DCM (ICM). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were used for differentially expressed genes (DEGs). The miRNA-mRNA interactions were identified by Pearson correlation analysis and miRNA target-prediction programs. mRNA-seq and miRNA-seq were validated by qRT-PCR and miRNA-mRNA interactions were validated by luciferase assays. We found 112 mRNAs and five miRNAs dysregulated in VCM vs. ICM. DEGs were positively enriched for pathways related to the extracellular matrix (ECM), mitochondrial respiration, cardiac muscle contraction, and fatty acid metabolism in VCM vs. ICM and negatively enriched for immune-response-related pathways, JAK-STAT, and NF-kappa B signaling. We identified four pairs of negatively correlated miRNA-mRNA: miR-218-5p-DDX6, miR-218-5p-TTC39C, miR-218-5p-SEMA4A, and miR-494-3p-SGMS2. Our study revealed novel miRNA-mRNA interaction networks and signaling pathways for VCM and ICM, providing novel insights into the development of these DCM etiologies.
Keywords: RNA sequencing; dilated cardiomyopathy; etiology; ischemic cardiomyopathy; microRNA; volume overload.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Kittleson M.M., Ye S.Q., Irizarry R.A., Minhas K.M., Edness G., Conte J.V., Parmigiani G., Miller L.W., Chen Y., Hall J.L., et al. Identification of a Gene Expression Profile That Differentiates between Ischemic and Nonischemic Cardiomyopathy. Circulation. 2004;110:3444–3451. doi: 10.1161/01.CIR.0000148178.19465.11. - DOI - PubMed
-
- Merlo M., Gentile P., Artico J., Cannatà A., Paldino A., De Angelis G., Barbati G., Alonge M., Gigli M., Pinamonti B., et al. Arrhythmic Risk Stratification in Patients with Dilated Cardiomyopathy and Intermediate Left Ventricular Dysfunction. J. Cardiovasc. Med. 2019;20:343–350. doi: 10.2459/JCM.0000000000000792. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

