Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction
- PMID: 38785939
- PMCID: PMC11117998
- DOI: 10.3390/biom14050532
Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction
Abstract
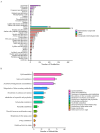
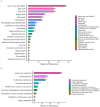
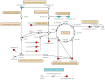
Myocardial infarction (MI), including ST-segment elevation MI (STEMI) and non-ST-segment elevation MI (NSTEMI), is still a leading cause of death worldwide. Metabolomics technology was used to explore differential metabolites (DMs) as potential biomarkers for early diagnosis of STEMI and NSTEMI. In the study, 2531 metabolites, including 1925 DMs, were discovered. In the selected 27 DMs, 14 were successfully verified in a new cohort, and the AUC values were all above 0.8. There were 10 in STEMI group, namely L-aspartic acid, L-acetylcarnitine, acetylglycine, decanoylcarnitine, hydroxyphenyllactic acid, ferulic acid, itaconic acid, lauroylcarnitine, myristoylcarnitine, and cis-4-hydroxy-D-proline, and 5 in NSTEMI group, namely L-aspartic acid, arachidonic acid, palmitoleic acid, D-aspartic acid, and palmitelaidic acid. These 14 DMs may be developed as biomarkers for the early diagnosis of MI with high sensitivity and specificity. These findings have particularly important clinical significance for NSTEMI patients because these patients have no typical ECG changes.
Keywords: NSTEMI; STEMI; acute coronary syndrome; biomarker; metabolomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
-
- American Heart Association Focus on Quality. [(accessed on 28 April 2023)]. Available online: http://www.heart.org/en/professional/quality-improvement.
-
- Tsao C.W., Aday A.W., Almarzooq Z.I., Anderson C.A., Arora P., Avery C.L., Baker-Smith C.M., Beaton A.Z., Boehme A.K., Buxton A.E., et al. Heart disease and stroke statistics-2023 update: A report from the American Heart Association. Circulation. 2023;147:e93–e621. doi: 10.1161/CIR.0000000000001123. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 82170353 & 82370350/National Natural Science Foundation of China
- 22ZYQYSY00020/Tianjin Municipal Science and Technology Commission
- 2020-PT310-007/Chinese Academy of Medical Sciences
- 2019-TD-003/TEDA International Cardiovascular Hospital
- TJYXZDXK-019A/Tianjin Key Medical Discipline (Specialty) Construction Project
LinkOut - more resources
Full Text Sources
Medical

