Generation of Rhesus Macaque Embryos with Expanded CAG Trinucleotide Repeats in the Huntingtin Gene
- PMID: 38786052
- PMCID: PMC11119628
- DOI: 10.3390/cells13100829
Generation of Rhesus Macaque Embryos with Expanded CAG Trinucleotide Repeats in the Huntingtin Gene
Abstract
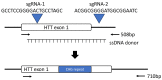
Huntington's disease (HD) arises from expanded CAG repeats in exon 1 of the Huntingtin (HTT) gene. The resultant misfolded HTT protein accumulates within neuronal cells, negatively impacting their function and survival. Ultimately, HTT accumulation results in cell death, causing the development of HD. A nonhuman primate (NHP) HD model would provide important insight into disease development and the generation of novel therapies due to their genetic and physiological similarity to humans. For this purpose, we tested CRISPR/Cas9 and a single-stranded DNA (ssDNA) containing expanded CAG repeats in introducing an expanded CAG repeat into the HTT gene in rhesus macaque embryos. Analyses were conducted on arrested embryos and trophectoderm (TE) cells biopsied from blastocysts to assess the insertion of the ssDNA into the HTT gene. Genotyping results demonstrated that 15% of the embryos carried an expanded CAG repeat. The integration of an expanded CAG repeat region was successfully identified in five blastocysts, which were cryopreserved for NHP HD animal production. Some off-target events were observed in biopsies from the cryopreserved blastocysts. NHP embryos were successfully produced, which will help to establish an NHP HD model and, ultimately, may serve as a vital tool for better understanding HD's pathology and developing novel treatments.
Keywords: CRISPR; Huntington’s disease; germline editing; rhesus macaque.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Generation of New Isogenic Models of Huntington's Disease Using CRISPR-Cas9 Technology.Int J Mol Sci. 2020 Mar 8;21(5):1854. doi: 10.3390/ijms21051854. Int J Mol Sci. 2020. PMID: 32182692 Free PMC article.
-
Towards a transgenic model of Huntington's disease in a non-human primate.Nature. 2008 Jun 12;453(7197):921-4. doi: 10.1038/nature06975. Epub 2008 May 18. Nature. 2008. PMID: 18488016 Free PMC article.
-
Pathogenic cellular phenotypes are germline transmissible in a transgenic primate model of Huntington's disease.Stem Cells Dev. 2013 Apr 15;22(8):1198-205. doi: 10.1089/scd.2012.0469. Epub 2013 Jan 15. Stem Cells Dev. 2013. PMID: 23190281 Free PMC article.
-
RNA toxicity induced by expanded CAG repeats in Huntington's disease.Brain Pathol. 2016 Nov;26(6):779-786. doi: 10.1111/bpa.12427. Brain Pathol. 2016. PMID: 27529325 Free PMC article. Review.
-
Huntington's Disease: Relationship Between Phenotype and Genotype.Mol Neurobiol. 2017 Jan;54(1):342-348. doi: 10.1007/s12035-015-9662-8. Epub 2016 Jan 7. Mol Neurobiol. 2017. PMID: 26742514 Review.
Cited by
-
Exploring immunotherapeutic strategies for neurodegenerative diseases: a focus on Huntington's disease and Prion diseases.Acta Pharmacol Sin. 2025 Jun;46(6):1511-1538. doi: 10.1038/s41401-024-01455-w. Epub 2025 Jan 31. Acta Pharmacol Sin. 2025. PMID: 39890942 Review.
-
Non-human primates as a translational model for the study of male reproductive health.Nat Rev Urol. 2025 Jul 7:10.1038/s41585-025-01062-2. doi: 10.1038/s41585-025-01062-2. Online ahead of print. Nat Rev Urol. 2025. PMID: 40624316 Review.
-
Non-human primate: the new frontier model of female reproductive engineering.Front Bioeng Biotechnol. 2025 Mar 28;13:1536750. doi: 10.3389/fbioe.2025.1536750. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40242357 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

