Uncovering miRNA-mRNA Regulatory Networks Related to Olaparib Resistance and Resensitization of BRCA2MUT Ovarian Cancer PEO1-OR Cells with the ATR/CHK1 Pathway Inhibitors
- PMID: 38786089
- PMCID: PMC11119970
- DOI: 10.3390/cells13100867
Uncovering miRNA-mRNA Regulatory Networks Related to Olaparib Resistance and Resensitization of BRCA2MUT Ovarian Cancer PEO1-OR Cells with the ATR/CHK1 Pathway Inhibitors
Abstract
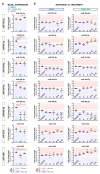
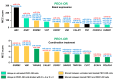
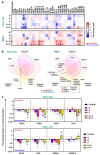
Resistance to olaparib is the major obstacle in targeted therapy for ovarian cancer (OC) with poly(ADP-ribose) polymerase inhibitors (PARPis), prompting studies on novel combination therapies to enhance olaparib efficacy. Despite identifying various mechanisms, understanding how OC cells acquire PARPi resistance remains incomplete. This study investigated microRNA (miRNA) expression in olaparib-sensitive (PEO1, PEO4) and previously established olaparib-resistant OC cell lines (PEO1-OR) using high-throughput RT-qPCR and bioinformatic analyses. The role of miRNAs was explored regarding acquired resistance and resensitization with the ATR/CHK1 pathway inhibitors. Differentially expressed miRNAs were used to construct miRNA-mRNA regulatory networks and perform functional enrichment analyses for target genes with miRNet 2.0. TCGA-OV dataset was analyzed to explore the prognostic value of selected miRNAs and target genes in clinical samples. We identified potential processes associated with olaparib resistance, including cell proliferation, migration, cell cycle, and growth factor signaling. Resensitized PEO1-OR cells were enriched in growth factor signaling via PDGF, EGFR, FGFR1, VEGFR2, and TGFβR, regulation of the cell cycle via the G2/M checkpoint, and caspase-mediated apoptosis. Antibody microarray analysis confirmed dysregulated growth factor expression. The addition of the ATR/CHK1 pathway inhibitors to olaparib downregulated FGF4, FGF6, NT-4, PLGF, and TGFβ1 exclusively in PEO1-OR cells. Survival and differential expression analyses for serous OC patients revealed prognostic miRNAs likely associated with olaparib resistance (miR-99b-5p, miR-424-3p, and miR-505-5p) and resensitization to olaparib (miR-324-5p and miR-424-3p). Essential miRNA-mRNA interactions were reconstructed based on prognostic miRNAs and target genes. In conclusion, our data highlight distinct miRNA profiles in olaparib-sensitive and olaparib-resistant cells, offering molecular insights into overcoming resistance with the ATR/CHK1 inhibitors in OC. Moreover, some miRNAs might serve as potential predictive signature molecules of resistance and therapeutic response.
Keywords: ATR/CHK1 pathway; TCGA data; bioinformatics; combination therapy; growth factors; miRNA profiling; olaparib; ovarian cancer; resistance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- DiSilvestro P., Colombo N., Scambia G., Kim B.G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. Efficacy of Maintenance Olaparib for Patients with Newly Diagnosed Advanced Ovarian Cancer with a BRCA Mutation: Subgroup Analysis Findings from the SOLO1 Trial. J. Clin. Oncol. 2020;38:3528–3537. doi: 10.1200/JCO.20.00799. - DOI - PMC - PubMed
-
- Nicum S., McGregor N., Austin R., Collins L., Dutton S., McNeish I., Glasspool R., Hall M., Roux R., Michael A., et al. Results of a randomised Phase II trial of olaparib, chemotherapy or olaparib and cediranib in patients with platinum-resistant ovarian cancer. Br. J. Cancer. 2024;130:941–950. doi: 10.1038/s41416-023-02567-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

