The MraY Inhibitor Muraymycin D2 and Its Derivatives Induce Enlarged Cells in Obligate Intracellular Chlamydia and Wolbachia and Break the Persistence Phenotype in Chlamydia
- PMID: 38786149
- PMCID: PMC11117252
- DOI: 10.3390/antibiotics13050421
The MraY Inhibitor Muraymycin D2 and Its Derivatives Induce Enlarged Cells in Obligate Intracellular Chlamydia and Wolbachia and Break the Persistence Phenotype in Chlamydia
Abstract
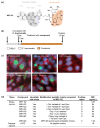
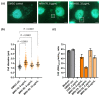
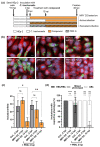
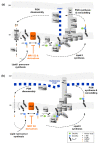
Chlamydial infections and diseases caused by filarial nematodes are global health concerns. However, treatment presents challenges due to treatment failures potentially caused by persisting Chlamydia and long regimens against filarial infections accompanied by low compliance. A new treatment strategy could be the targeting of the reduced peptidoglycan structures involved in cell division in the obligate intracellular bacteria Chlamydia and Wolbachia, the latter being obligate endosymbionts supporting filarial development, growth, and survival. Here, cell culture experiments with C. trachomatis and Wolbachia showed that the nucleoside antibiotics muraymycin and carbacaprazamycin interfere with bacterial cell division and induce enlarged, aberrant cells resembling the penicillin-induced persistence phenotype in Chlamydia. Enzymatic inhibition experiments with purified C. pneumoniae MraY revealed that muraymycin derivatives abolish the synthesis of the peptidoglycan precursor lipid I. Comparative in silico analyses of chlamydial and wolbachial MraY with the corresponding well-characterized enzyme in Aquifex aeolicus revealed a high degree of conservation, providing evidence for a similar mode of inhibition. Muraymycin D2 treatment eradicated persisting non-dividing C. trachomatis cells from an established penicillin-induced persistent infection. This finding indicates that nucleoside antibiotics may have additional properties that can break bacterial persistence.
Keywords: Chlamydia; MraY; Wolbachia; cell division; intracellular bacteria; lipid II synthesis; muraymycin; peptidoglycan; persistence-breaking.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- European Centre for Disease Prevention and Control . ECDC. Annual Epidemiological Report for 2019. ECDC; Stockholm, Switzerland: 2022. Chlamydia Infection.
-
- Weström L., Joesoef R., Reynolds G., Hagdu A., Thompson S.E. Pelvic Inflammatory Disease and Fertility: A Cohort Study of 1844 Women with Laparoscopically Verified Disease and 657 Control Women with Normal Laparoscopic Results. Sex. Transm. Dis. 1992;19:185–192. doi: 10.1097/00007435-199207000-00001. - DOI - PubMed
-
- WHO . WHO Guidelines for the Treatment of Chlamydia Trachomatis. World Health Organization; Geneva, Switzerland: 2016. - PubMed
Grants and funding
- 398967434 - TRR261/Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)
- N.a./BONFOR intramural funding program, Medical Faculty of Bonn University
- N.a./Jürgen Manchot foundation
- N.a./FEMHABIL, Medical Faculty, University of Bonn
- JP22H02738/JSPS KAKENHI Grant-in-Aid for Scientific Research (B)
LinkOut - more resources
Full Text Sources
Medical

